Cell culture-adaptive mutations in hepatitis C virus promote viral production by enhancing viral replication and release
- PMID: 29599605
- PMCID: PMC5871825
- DOI: 10.3748/wjg.v24.i12.1299
Cell culture-adaptive mutations in hepatitis C virus promote viral production by enhancing viral replication and release
Abstract
Aim: To explore hepatitis C virus (HCV) adaptive mutations or combinations thereof responsible for enhanced viral production and investigate the underlying mechanisms.
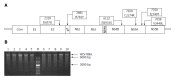
Methods: A series of plasmids with adaptive mutations were constructed. After the plasmids were transfected into Huh7.5 cells, we determined the infectious HCV particle titers by NS5A immunofluorescence assays, and detected HCV RNA replication by real-time PCR and protein expression by Western blot. Then we carried out immunoblotting of supernatants and cell lysates with anti-NS3 to analyze the virus release level. In addition, co-localization of lipid droplets (LDs) with NS5A was measured using confocal laser scanning microscopy. The ratio between the p56 and p58 phosphoforms of NS5A was analyzed further.
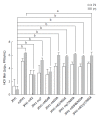
Results: The plasmids named JFH1-mE2, JFH1-mp7, JFH1-mNS4B, JFH1-mNS5A, JFH1-mE2/NS5A, JFH1-mp7/NS5A, JFH1-mNS4B/NS5A, JFH1-mE2/p7/NS5A, and mJFH1 were constructed successfully. This study generated infectious HCV particles with a robust titer of 1.61 × 106 focus-forming units (FFUs)/mL. All of the six adaptive mutations increased the HCV particle production at varying levels. The NS5A (C2274R, I2340T, and V2440L) and p7 (H781Y) were critical adaptive mutations. The effect of NS5A (C2274R, I2340T, and V2440L), p7 (H781Y), and NS4B (N1931S) on infectious HCV titers was investigated by measuring the HCV RNA replication, protein expression, and virion release. However, the six adaptive mutations were not required for the LD localization of NS5A proteins or the phosphorylation of NS5A.
Conclusion: In this study, we generated infectious HCV particles with a robust titer of 1.61 × 106 FFUs/mL, and found that the viral replication and release levels could be enhanced by some of the adaptive mutations.
Keywords: Adaptive mutation; Hepatitis C virus; JFH1; Lipid droplet localization; RNA replication; Virion release.
Conflict of interest statement
Conflict-of-interest statement: The authors declare that there are no conflicts of interest in this study.
Figures
Similar articles
-
A cell culture adapted HCV JFH1 variant that increases viral titers and permits the production of high titer infectious chimeric reporter viruses.PLoS One. 2012;7(9):e44965. doi: 10.1371/journal.pone.0044965. Epub 2012 Sep 13. PLoS One. 2012. PMID: 23028707 Free PMC article.
-
Adaptive Mutations Enhance Assembly and Cell-to-Cell Transmission of a High-Titer Hepatitis C Virus Genotype 5a Core-NS2 JFH1-Based Recombinant.J Virol. 2015 Aug;89(15):7758-75. doi: 10.1128/JVI.00039-15. Epub 2015 May 20. J Virol. 2015. PMID: 25995244 Free PMC article.
-
Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses.Gastroenterology. 2007 Nov;133(5):1614-26. doi: 10.1053/j.gastro.2007.08.005. Epub 2007 Aug 3. Gastroenterology. 2007. PMID: 17983807
-
[Involvement of nonstructural protein 5A and lipids on production of hepatitis C virus particles].Uirusu. 2008 Dec;58(2):199-205. doi: 10.2222/jsv.58.199. Uirusu. 2008. PMID: 19374198 Review. Japanese.
-
Current status and future development of infectious cell-culture models for the major genotypes of hepatitis C virus: Essential tools in testing of antivirals and emerging vaccine strategies.Antiviral Res. 2018 Oct;158:264-287. doi: 10.1016/j.antiviral.2018.07.014. Epub 2018 Jul 27. Antiviral Res. 2018. PMID: 30059723 Review.
Cited by
-
Progress, evolving therapeutic/diagnostic approaches, and challenges in the management of hepatitis C virus infections.Arch Virol. 2022 Mar;167(3):717-736. doi: 10.1007/s00705-022-05375-0. Epub 2022 Jan 28. Arch Virol. 2022. PMID: 35089390 Free PMC article. Review.
-
Unraveling the dynamics of hepatitis C virus adaptive mutations and their impact on antiviral responses in primary human hepatocytes.J Virol. 2024 Mar 19;98(3):e0192123. doi: 10.1128/jvi.01921-23. Epub 2024 Feb 6. J Virol. 2024. PMID: 38319104 Free PMC article.
-
Relationship between hepatitis C and kidney stone in US females: Results from the National Health and Nutrition Examination Survey in 2007-2018.Front Public Health. 2022 Aug 5;10:940905. doi: 10.3389/fpubh.2022.940905. eCollection 2022. Front Public Health. 2022. PMID: 35991057 Free PMC article.
References
-
- Averhoff FM, Glass N, Holtzman D. Global burden of hepatitis C: considerations for healthcare providers in the United States. Clin Infect Dis. 2012;55 Suppl 1:S10–S15. - PubMed
-
- Hoofnagle JH, di Bisceglie AM. The treatment of chronic viral hepatitis. N Engl J Med. 1997;336:347–356. - PubMed
-
- World Health Organization. Guidelines for the Screening Care and Treatment of Persons with Chronic Hepatitis C Infection: Updated Version. Geneva: World Health Organization;; 2016. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

