Transcriptional Response of Staphylococcus aureus to Sunlight in Oxic and Anoxic Conditions
- PMID: 29599752
- PMCID: PMC5863498
- DOI: 10.3389/fmicb.2018.00249
Transcriptional Response of Staphylococcus aureus to Sunlight in Oxic and Anoxic Conditions
Abstract
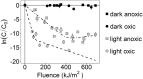
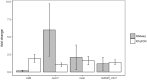
The transcriptional response of Staphylococcus aureus strain Newman to sunlight exposure was investigated under both oxic and anoxic conditions using RNA sequencing to gain insight into potential mechanisms of inactivation. S. aureus is a pathogenic bacterium detected at recreational beaches which can cause gastrointestinal illness and skin infections, and is of increasing public health concern. To investigate the S. aureus photostress response in oligotrophic seawater, S. aureus cultures were suspended in seawater and exposed to full spectrum simulated sunlight. Experiments were performed under oxic or anoxic conditions to gain insight into the effects of oxygen-mediated and non-oxygen-mediated inactivation mechanisms. Transcript abundance was measured after 6 h of sunlight exposure using RNA sequencing and was compared to transcript abundance in paired dark control experiments. Culturable S. aureus decayed following biphasic inactivation kinetics with initial decay rate constants of 0.1 and 0.03 m2 kJ-1 in oxic and anoxic conditions, respectively. RNA sequencing revealed that 71 genes had different transcript abundance in the oxic sunlit experiments compared to dark controls, and 18 genes had different transcript abundance in the anoxic sunlit experiments compared to dark controls. The majority of genes showed reduced transcript abundance in the sunlit experiments under both conditions. Three genes (ebpS, NWMN_0867, and NWMN_1608) were found to have the same transcriptional response to sunlight between both oxic and anoxic conditions. In the oxic condition, transcripts associated with porphyrin metabolism, nitrate metabolism, and membrane transport functions were increased in abundance during sunlight exposure. Results suggest that S. aureus responds differently to oxygen-dependent and oxygen-independent photostress, and that endogenous photosensitizers play an important role during oxygen-dependent indirect photoinactivation.
Keywords: RNA; Staphylococcus; photoinactivation; sequencing; sunlight; transcription.
Figures


References
-
- Al-Jassim N., Mantilla-Calderon D., Wang T., Hong P.-Y. (2017). Inactivation of a virulent wastewater Escherichia coli and non-virulent commensal Escherichia coli DSM1103 strains and their gene expression upon solar irradiation. Environ. Sci. Technol. 51, 3649–3659. 10.1021/acs.est.6b05377 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

