An Updated Functional Annotation of Protein-Coding Genes in the Cucumber Genome
- PMID: 29599790
- PMCID: PMC5863696
- DOI: 10.3389/fpls.2018.00325
An Updated Functional Annotation of Protein-Coding Genes in the Cucumber Genome
Abstract
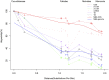
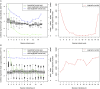
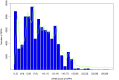
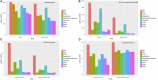
Background: Although the cucumber reference genome and its annotation were published several years ago, the functional annotation of predicted genes, particularly protein-coding genes, still requires further improvement. In general, accurately determining orthologous relationships between genes allows for better and more robust functional assignments of predicted genes. As one of the most reliable strategies, the determination of collinearity information may facilitate reliable orthology inferences among genes from multiple related genomes. Currently, the identification of collinear segments has mainly been based on conservation of gene order and orientation. Over the course of plant genome evolution, various evolutionary events have disrupted or distorted the order of genes along chromosomes, making it difficult to use those genes as genome-wide markers for plant genome comparisons. Results: Using the localized LASTZ/MULTIZ analysis pipeline, we aligned 15 genomes, including cucumber and other related angiosperm plants, and identified a set of genomic segments that are short in length, stable in structure, uniform in distribution and highly conserved across all 15 plants. Compared with protein-coding genes, these conserved segments were more suitable for use as genomic markers for detecting collinear segments among distantly divergent plants. Guided by this set of identified collinear genomic segments, we inferred 94,486 orthologous protein-coding gene pairs (OPPs) between cucumber and 14 other angiosperm species, which were used as proxies for transferring functional terms to cucumber genes from the annotations of the other 14 genomes. In total, 10,885 protein-coding genes were assigned Gene Ontology (GO) terms which was nearly 1,300 more than results collected in Uniprot-proteomic database. Our results showed that annotation accuracy would been improved compared with other existing approaches. Conclusions: In this study, we provided an alternative resource for the functional annotation of predicted cucumber protein-coding genes, which we expect will be beneficial for the cucumber's biological study, accessible from http://cmb.bnu.edu.cn/functional_annotation. Meanwhile, using the cucumber reference genome as a case study, we presented an efficient strategy for transferring gene functional information from previously well-characterized protein-coding genes in model species to newly sequenced or "non-model" plant species.
Keywords: collinear segments; cucumber; gene functional annotation; orthology; protein-coding gene.
Figures




Similar articles
-
RNA-Seq improves annotation of protein-coding genes in the cucumber genome.BMC Genomics. 2011 Nov 2;12:540. doi: 10.1186/1471-2164-12-540. BMC Genomics. 2011. PMID: 22047402 Free PMC article.
-
[Analysis, identification and correction of some errors of model refseqs appeared in NCBI Human Gene Database by in silico cloning and experimental verification of novel human genes].Yi Chuan Xue Bao. 2004 May;31(5):431-43. Yi Chuan Xue Bao. 2004. PMID: 15478601 Chinese.
-
Conservation and functional element discovery in 20 angiosperm plant genomes.Mol Biol Evol. 2013 Jul;30(7):1729-44. doi: 10.1093/molbev/mst082. Epub 2013 May 2. Mol Biol Evol. 2013. PMID: 23640124
-
Inferring Orthologs: Open Questions and Perspectives.Genomics Insights. 2016 Feb 25;9:17-28. doi: 10.4137/GEI.S37925. eCollection 2016. Genomics Insights. 2016. PMID: 26966373 Free PMC article. Review.
-
Genome-wide functional annotation of variants: a systematic review of state-of-the-art tools, techniques and resources.Front Pharmacol. 2025 Mar 3;16:1474026. doi: 10.3389/fphar.2025.1474026. eCollection 2025. Front Pharmacol. 2025. PMID: 40098614 Free PMC article. Review.
Cited by
-
Identification of clade-wide putative cis-regulatory elements from conserved non-coding sequences in Cucurbitaceae genomes.Hortic Res. 2023 Feb 28;10(4):uhad038. doi: 10.1093/hr/uhad038. eCollection 2023 Apr. Hortic Res. 2023. PMID: 37799630 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

