Human muscle-specific A-kinase anchoring protein polymorphisms modulate the susceptibility to cardiovascular diseases by altering cAMP/PKA signaling
- PMID: 29600899
- PMCID: PMC6087776
- DOI: 10.1152/ajpheart.00034.2018
Human muscle-specific A-kinase anchoring protein polymorphisms modulate the susceptibility to cardiovascular diseases by altering cAMP/PKA signaling
Erratum in
-
Corrigendum.Am J Physiol Heart Circ Physiol. 2019 Feb 1;316(2):H432. doi: 10.1152/ajpheart.zh4-2504-corr.2019. Am J Physiol Heart Circ Physiol. 2019. PMID: 30715904 Free PMC article. No abstract available.
Abstract
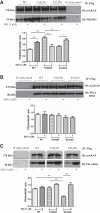
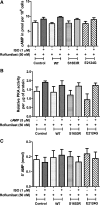
One of the crucial cardiac signaling pathways is cAMP-mediated PKA signal transduction, which is regulated by a family of scaffolding proteins, i.e., A-kinase anchoring proteins (AKAPs). Muscle-specific AKAP (mAKAP) partly regulates cardiac cAMP/PKA signaling by binding to PKA and phosphodiesterase 4D3 (PDE4D3), among other proteins, and plays a central role in modulating cardiac remodeling. Moreover, genetics plays an incomparable role in modifying the risk of cardiovascular diseases (CVDs). Single-nucleotide polymorphisms (SNPs) in various proteins have especially been shown to predispose individuals to CVDs. Hence, we hypothesized that human mAKAP polymorphisms found in humans with CVDs alter the cAMP/PKA pathway, influencing the susceptibility of individuals to CVDs. Our computational analyses revealed two mAKAP SNPs found in cardiac disease-related patients with the highest predicted deleterious effects, Ser 1653 Arg (S1653R) and Glu 2124 Gly (E2124G). Coimmunoprecipitation data in human embryonic kidney-293T cells showed that the S1653R SNP, present in the PDE4D3-binding domain of mAKAP, changed the binding of PDE4D3 to mAKAP and that the E2124G SNP, flanking the 3'-PKA binding domain, changed the binding of PKA before and after stimulation with isoproterenol. These SNPs significantly altered intracellular cAMP levels, global PKA activity, and cytosolic PDE activity compared with the wild type before and after isoproterenol stimulation. PKA-mediated phosphorylation of pathological markers was found to be upregulated after cell stimulation in both mutants. In conclusion, human mAKAP polymorphisms may influence the propensity of developing CVDs by affecting cAMP/PKA signaling, supporting the clinical significance of PKA-mAKAP-PDE4D3 interactions. NEW & NOTEWORTHY We found that single-nucleotide polymorphisms in muscle-specific A-kinase anchoring protein found in human patients with cardiovascular diseases significantly affect the cAMP/PKA signaling pathway. Our results showed, for the first time, that human muscle-specific A-kinase anchoring protein polymorphisms might alter the susceptibility of individuals to develop cardiovascular diseases with known underlying molecular mechanisms.
Figures





Comment in
-
Form follows function: polymorphisms in mAKAP alter cardiac cAMP/PKA signaling.Am J Physiol Heart Circ Physiol. 2018 Sep 1;315(3):H626-H628. doi: 10.1152/ajpheart.00248.2018. Epub 2018 May 4. Am J Physiol Heart Circ Physiol. 2018. PMID: 29727216 Free PMC article. No abstract available.
References
-
- Bidulescu A, Liu J, Musani SK, Fox ER, Samdarshi TE, Sarpong DF, Vaccarino V, Wilson PW, Arnett DK, Din-Dzietham R, Taylor HA, Gibbons GH. Association of adiponectin with left ventricular mass in blacks: the Jackson Heart Study. Circ Heart Fail 4: 747–753, 2011. doi: 10.1161/CIRCHEARTFAILURE.110.959742. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

