Classical and alternative roles for autophagy in lipid metabolism
- PMID: 29601311
- PMCID: PMC5930069
- DOI: 10.1097/MOL.0000000000000509
Classical and alternative roles for autophagy in lipid metabolism
Abstract
Purpose of review: Intracellular lipid metabolism is a complex interplay of exogenous lipid handling, trafficking, storage, lipolysis, and export. Recent work has implicated the cellular degradative process called autophagy in several aspects of lipid metabolism. We will discuss both the classical and novel roles of autophagy and the autophagic machinery in this setting.
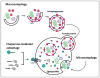
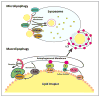
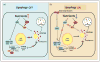
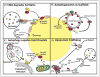
Recent findings: The delivery of lipid droplets to lysosomes for hydrolysis, named lipophagy, was the first described functional role for autophagy in lipid metabolism. The molecular machinery and regulation of this selective form of macroautophagy is beginning to be discovered and has the potential to shed enormous light on intracellular lipolysis. Yet, the autophagic machinery appears to also be coopted for alternative roles that include interaction with cytosolic lipolysis pathways, supply and expansion of lipid droplets, and lipoprotein trafficking. Additionally, lesser studied forms of autophagy called microautophagy and chaperone-mediated autophagy have distinct roles in lipid handling that also intersect with classical macroautophagy. The integration of current knowledge in these areas into a holistic understanding of intracellular lipid metabolism will be a goal of this review.
Summary: As the field of autophagy has evolved and expanded to include functional roles in various aspects of cellular degradation, so has its role in intracellular lipid metabolism. Understanding the mechanisms underlying these classical and alternative roles of autophagy will not only enhance our knowledge in lipid biology but also provide new avenues of translation to human lipid disorders.
Conflict of interest statement
There are no conflicts of interest.
Figures
Similar articles
-
Mechanisms of lipid droplet degradation.Curr Opin Cell Biol. 2024 Oct;90:102402. doi: 10.1016/j.ceb.2024.102402. Epub 2024 Jul 24. Curr Opin Cell Biol. 2024. PMID: 39053179 Review.
-
Lipid metabolism and lipophagy in cancer.Biochem Biophys Res Commun. 2018 Oct 7;504(3):582-589. doi: 10.1016/j.bbrc.2018.02.097. Epub 2018 Feb 10. Biochem Biophys Res Commun. 2018. PMID: 29438712 Free PMC article. Review.
-
Regulation and Functions of Autophagic Lipolysis.Trends Endocrinol Metab. 2016 Oct;27(10):696-705. doi: 10.1016/j.tem.2016.06.003. Epub 2016 Jun 27. Trends Endocrinol Metab. 2016. PMID: 27365163 Free PMC article. Review.
-
Lipophagy and liver disease: New perspectives to better understanding and therapy.Biomed Pharmacother. 2018 Jan;97:339-348. doi: 10.1016/j.biopha.2017.07.168. Epub 2017 Nov 6. Biomed Pharmacother. 2018. PMID: 29091883 Review.
-
Lipid Droplets and Their Autophagic Turnover via the Raft-Like Vacuolar Microdomains.Int J Mol Sci. 2021 Jul 29;22(15):8144. doi: 10.3390/ijms22158144. Int J Mol Sci. 2021. PMID: 34360917 Free PMC article. Review.
Cited by
-
The role of autophagy/lipophagy in the response of osteoblastic cells to hyperlipidemia (Review).Exp Ther Med. 2024 Jun 19;28(2):328. doi: 10.3892/etm.2024.12617. eCollection 2024 Aug. Exp Ther Med. 2024. PMID: 38979020 Free PMC article. Review.
-
Catalpol induces autophagy and attenuates liver steatosis in ob/ob and high-fat diet-induced obese mice.Aging (Albany NY). 2019 Nov 7;11(21):9461-9477. doi: 10.18632/aging.102396. Epub 2019 Nov 7. Aging (Albany NY). 2019. PMID: 31697646 Free PMC article.
-
Control of NAD+ homeostasis by autophagic flux modulates mitochondrial and cardiac function.EMBO J. 2024 Feb;43(3):362-390. doi: 10.1038/s44318-023-00009-w. Epub 2024 Jan 11. EMBO J. 2024. PMID: 38212381 Free PMC article.
-
Metabolic Reprogramming of Macrophages in Atherosclerosis: Is It All about Cholesterol?J Lipid Atheroscler. 2020 May;9(2):231-242. doi: 10.12997/jla.2020.9.2.231. Epub 2020 Mar 3. J Lipid Atheroscler. 2020. PMID: 32821733 Free PMC article. Review.
-
Ginsenoside Compound K Protects against Obesity through Pharmacological Targeting of Glucocorticoid Receptor to Activate Lipophagy and Lipid Metabolism.Pharmaceutics. 2022 Jun 2;14(6):1192. doi: 10.3390/pharmaceutics14061192. Pharmaceutics. 2022. PMID: 35745765 Free PMC article.
References
-
- Arner P, Langin D. Lipolysis in lipid turnover, cancer cachexia, and obesity-induced insulin resistance. Trends Endocrinol Metab. 2014;25:255–262. - PubMed
-
- Subramanian V, Rothenberg A, Gomez C, et al. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem. 2004;279:42062–42071. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

