Invertebrate Iridoviruses: A Glance over the Last Decade
- PMID: 29601483
- PMCID: PMC5923455
- DOI: 10.3390/v10040161
Invertebrate Iridoviruses: A Glance over the Last Decade
Abstract
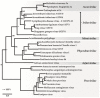
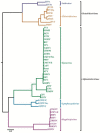
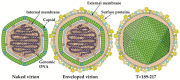
Members of the family Iridoviridae (iridovirids) are large dsDNA viruses that infect both invertebrate and vertebrate ectotherms and whose symptoms range in severity from minor reductions in host fitness to systemic disease and large-scale mortality. Several characteristics have been useful for classifying iridoviruses; however, novel strains are continuously being discovered and, in many cases, reliable classification has been challenging. Further impeding classification, invertebrate iridoviruses (IIVs) can occasionally infect vertebrates; thus, host range is often not a useful criterion for classification. In this review, we discuss the current classification of iridovirids, focusing on genomic and structural features that distinguish vertebrate and invertebrate iridovirids and viral factors linked to host interactions in IIV6 (Invertebrate iridescent virus 6). In addition, we show for the first time how complete genome sequences of viral isolates can be leveraged to improve classification of new iridovirid isolates and resolve ambiguous relations. Improved classification of the iridoviruses may facilitate the identification of genus-specific virulence factors linked with diverse host phenotypes and host interactions.
Keywords: classification; genomics; invertebrate iridoviruses; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Chinchar V., Essbauer S., He J., Hyatt A., Miyazaki T., Seligy V., Williams T. Virus Taxonomy Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; San Diego, CA, USA: 2005. Family Iridoviridae; pp. 145–162.
-
- King A.M., Lefkowitz E., Adams M.J., Carstens E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; Amsterdam, The Netherlands: 2011.
-
- Xeros N. A second virus disease of the leatherjacket, Tipula paludosa. Nature. 1954;174:562–563. doi: 10.1038/174562a0. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

