2'-O-methylation of the mRNA cap protects RNAs from decapping and degradation by DXO
- PMID: 29601584
- PMCID: PMC5877831
- DOI: 10.1371/journal.pone.0193804
2'-O-methylation of the mRNA cap protects RNAs from decapping and degradation by DXO
Erratum in
-
Correction: 2'-O-methylation of the mRNA cap protects RNAs from decapping and degradation by DXO.PLoS One. 2018 Aug 9;13(8):e0202308. doi: 10.1371/journal.pone.0202308. eCollection 2018. PLoS One. 2018. PMID: 30092053 Free PMC article.
Abstract
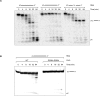
The 5' RNA cap structure (m7GpppRNA) is a key feature of eukaryotic mRNAs with important roles in stability, splicing, polyadenylation, mRNA export, and translation. Higher eukaryotes can further modify this minimal cap structure with the addition of a methyl group on the ribose 2'-O position of the first transcribed nucleotide (m7GpppNmpRNA) and sometimes on the adjoining nucleotide (m7GpppNmpNmpRNA). In higher eukaryotes, the DXO protein was previously shown to be responsible for both decapping and degradation of RNA transcripts harboring aberrant 5' ends such as pRNA, pppRNA, GpppRNA, and surprisingly, m7GpppRNA. It was proposed that the interaction of the cap binding complex with the methylated cap would prevent degradation of m7GpppRNAs by DXO. However, the critical role of the 2'-O-methylation found in higher eukaryotic cap structures was not previously addressed. In the present study, we demonstrate that DXO possesses both decapping and exoribonuclease activities toward incompletely capped RNAs, only sparing RNAs with a 2'-O-methylated cap structure. Fluorescence spectroscopy assays also revealed that the presence of the 2'-O-methylation on the cap structure drastically reduces the affinity of DXO for RNA. Moreover, immunofluorescence and structure-function assays also revealed that a nuclear localisation signal is located in the amino-terminus region of DXO. Overall, these results are consistent with a quality control mechanism in which DXO degrades incompletely capped RNAs.
Conflict of interest statement
Figures




References
-
- Ramanathan A, Robb GB, Chan S-H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44(16):7511–26. doi: 10.1093/nar/gkw551 - DOI - PMC - PubMed
-
- Wei C-M, Gershowitz A, Moss B. Methylated Nucleotides Block 5’ Terminus of HeLa Cell Messenger RNA. Cell. 1975;4(4):379–86. - PubMed
-
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, et al. 2’-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468(7322):452–6. doi: 10.1038/nature09489 - DOI - PMC - PubMed
-
- Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, et al. Ribose 2’-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12(2):137–43. doi: 10.1038/ni.1979 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

