The non-human primate kidney transcriptome in fetal development
- PMID: 29603257
- PMCID: PMC5963710
- DOI: 10.1111/jmp.12340
The non-human primate kidney transcriptome in fetal development
Abstract
Background: Little is known about the repertoire of non-human primate kidney genes expressed throughout development. The present work establishes an understanding of the primate renal transcriptome during fetal development in the context of renal maturation.
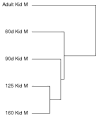
Methods: The baboon kidney transcriptome was characterized at 60-day gestation (DG), 90 DG, 125 DG, 140 DG, 160 DG and adulthood (6-12 years) using gene arrays and validated by QRT-PCR. Pathway and cluster analyses were used to characterize gene expression in the context of biological pathways.
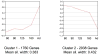
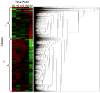
Results: Pathway analysis indicated activation of pathways not previously reported as relevant to kidney development. Cluster analysis also revealed gene splice variants with discordant expression profiles during development.
Conclusions: This study provides the first detailed genetic analysis of the developing primate kidney, and our findings of discordant expression of gene splice variants suggest that gene arrays likely provide a simplified view and demonstrate the need to study the fetal renal proteome.
Keywords: array; baboon; cluster analysis; gene expression; ontogeny; pathway analysis.
© 2018 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures
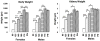


References
-
- Ingelfinger JR, Woods LL. Perinatal programming, renal development, and adult renal function. American Journal of Hypertension. 2002 Feb;15(2 Pt 2):46S–9S. - PubMed
-
- Rasch R, Skriver E, Woods LL. The role of the RAS in programming of adult hypertension. Acta Physiologica Scandinavica. 2004 Aug;181(4):537–42. - PubMed
-
- Schwab K, Patterson LT, Aronow BJ, Luckas R, Liang H-C, Potter SS. A catalogue of gene expression in the developing kidney. Kidney Int. 2003 Nov;64(5):1588–604. - PubMed
-
- McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, et al. GUDMAP: The genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008 Apr;19(4):667–71. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

