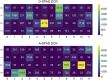
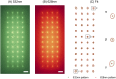
48-spot single-molecule FRET setup with periodic acceptor excitation
- PMID: 29604810
- PMCID: PMC5669981
- DOI: 10.1063/1.5000742
48-spot single-molecule FRET setup with periodic acceptor excitation
Abstract
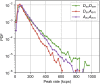
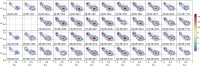
Single-molecule Förster resonance energy transfer (smFRET) allows measuring distances between donor and acceptor fluorophores on the 3-10 nm range. Solution-based smFRET allows measurement of binding-unbinding events or conformational changes of dye-labeled biomolecules without ensemble averaging and free from surface perturbations. When employing dual (or multi) laser excitation, smFRET allows resolving the number of fluorescent labels on each molecule, greatly enhancing the ability to study heterogeneous samples. A major drawback to solution-based smFRET is the low throughput, which renders repetitive measurements expensive and hinders the ability to study kinetic phenomena in real-time. Here we demonstrate a high-throughput smFRET system that multiplexes acquisition by using 48 excitation spots and two 48-pixel single-photon avalanche diode array detectors. The system employs two excitation lasers allowing separation of species with one or two active fluorophores. The performance of the system is demonstrated on a set of doubly labeled double-stranded DNA oligonucleotides with different distances between donor and acceptor dyes along the DNA duplex. We show that the acquisition time for accurate subpopulation identification is reduced from several minutes to seconds, opening the way to high-throughput screening applications and real-time kinetics studies of enzymatic reactions such as DNA transcription by bacterial RNA polymerase.
Figures


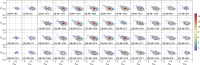
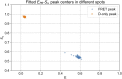

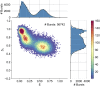


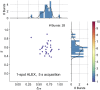


Similar articles
-
High-throughput smFRET analysis of freely diffusing nucleic acid molecules and associated proteins.Methods. 2019 Oct 1;169:21-45. doi: 10.1016/j.ymeth.2019.07.021. Epub 2019 Jul 26. Methods. 2019. PMID: 31356875 Free PMC article.
-
8-spot smFRET analysis using two 8-pixel SPAD arrays.Proc SPIE Int Soc Opt Eng. 2013 Feb 22;8590:10.1117/12.2003704. doi: 10.1117/12.2003704. Proc SPIE Int Soc Opt Eng. 2013. PMID: 24386541 Free PMC article.
-
Parallel multispot smFRET analysis using an 8-pixel SPAD array.Proc SPIE Int Soc Opt Eng. 2012 Jan 21;8228:10.1117/12.909470. doi: 10.1117/12.909470. Proc SPIE Int Soc Opt Eng. 2012. PMID: 24382989 Free PMC article.
-
Alternating-laser excitation: single-molecule FRET and beyond.Chem Soc Rev. 2014 Feb 21;43(4):1156-71. doi: 10.1039/c3cs60233h. Chem Soc Rev. 2014. PMID: 24037326 Review.
-
Multicolor single-molecule FRET for DNA and RNA processes.Curr Opin Struct Biol. 2021 Oct;70:26-33. doi: 10.1016/j.sbi.2021.03.005. Epub 2021 Apr 21. Curr Opin Struct Biol. 2021. PMID: 33894656 Free PMC article. Review.
Cited by
-
Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer.Science. 2018 Jan 19;359(6373):eaan1133. doi: 10.1126/science.aan1133. Science. 2018. PMID: 29348210 Free PMC article. Review.
-
Confinement-Free Wide-Field Ratiometric Tracking of Single Fluorescent Molecules.Biophys J. 2019 Dec 3;117(11):2141-2153. doi: 10.1016/j.bpj.2019.10.033. Epub 2019 Oct 31. Biophys J. 2019. PMID: 31711608 Free PMC article.
-
Real-Time Affinity Measurements of Proteins Synthesized in Cell-Free Lysate Using Fluorescence Correlation Spectroscopy.Anal Chem. 2025 May 13;97(18):9638-9647. doi: 10.1021/acs.analchem.4c05485. Epub 2025 Apr 30. Anal Chem. 2025. PMID: 40305650
-
Single-photon avalanche diode imagers in biophotonics: review and outlook.Light Sci Appl. 2019 Sep 18;8:87. doi: 10.1038/s41377-019-0191-5. eCollection 2019. Light Sci Appl. 2019. PMID: 31645931 Free PMC article. Review.
-
Integrated avalanche photodetectors for visible light.Nat Commun. 2021 Mar 23;12(1):1834. doi: 10.1038/s41467-021-22046-x. Nat Commun. 2021. PMID: 33758190 Free PMC article.
References
-
- Dahan M., Deniz A. A., Ha T., Chemla D. S., Schultz P. G., and Weiss S., Chem. Phys. 247, 85 (1999).10.1016/s0301-0104(99)00132-9 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

