Ensemble Docking in Drug Discovery
- PMID: 29606412
- PMCID: PMC6129458
- DOI: 10.1016/j.bpj.2018.02.038
Ensemble Docking in Drug Discovery
Abstract
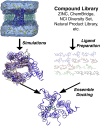
Ensemble docking corresponds to the generation of an "ensemble" of drug target conformations in computational structure-based drug discovery, often obtained by using molecular dynamics simulation, that is used in docking candidate ligands. This approach is now well established in the field of early-stage drug discovery. This review gives a historical account of the development of ensemble docking and discusses some pertinent methodological advances in conformational sampling.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Kaldor S.W., Kalish V.J., Tatlock J.H. Viracept (nelfinavir mesylate, AG1343): a potent, orally bioavailable inhibitor of HIV-1 protease. J. Med. Chem. 1997;40:3979–3985. - PubMed
-
- von Itzstein M., Wu W.Y., Oliver S.W. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. - PubMed
-
- Elber R., Karplus M. Multiple conformational states of proteins: a molecular dynamics analysis of myoglobin. Science. 1987;235:318–321. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

