Visualization of Transvection in Living Drosophila Embryos
- PMID: 29606591
- PMCID: PMC6092965
- DOI: 10.1016/j.molcel.2018.02.029
Visualization of Transvection in Living Drosophila Embryos
Abstract
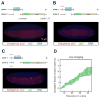
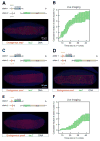
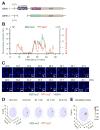
How remote enhancers interact with appropriate target genes persists as a central mystery in gene regulation. Here, we exploit the properties of transvection to explore enhancer-promoter communication between homologous chromosomes in living Drosophila embryos. We successfully visualized the activation of an MS2-tagged reporter gene by a defined developmental enhancer located in trans on the other homolog. This trans-homolog activation depends on insulator DNAs, which increase the stability-but not the frequency-of homolog pairing. A pair of heterotypic insulators failed to mediate transvection, raising the possibility that insulator specificity underlies the formation of chromosomal loop domains. Moreover, we found that a shared enhancer co-activates separate PP7 and MS2 reporter genes incis and intrans. Transvecting alleles weakly compete with one another, raising the possibility that they share a common pool of the transcription machinery. We propose that transvecting alleles form a trans-homolog "hub," which serves as a scaffold for the accumulation of transcription complexes.
Keywords: Drosophila embryos; TADs; chromosomal loop domains; enhancers; insulators; live imaging; transcription; transvection.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

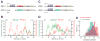


Comment in
-
Transvection Goes Live-Visualizing Enhancer-Promoter Communication between Chromosomes.Mol Cell. 2018 Apr 19;70(2):195-196. doi: 10.1016/j.molcel.2018.04.004. Mol Cell. 2018. PMID: 29677489
References
-
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

