Regulation of carbohydrate degradation pathways in Pseudomonas involves a versatile set of transcriptional regulators
- PMID: 29607620
- PMCID: PMC5902321
- DOI: 10.1111/1751-7915.13263
Regulation of carbohydrate degradation pathways in Pseudomonas involves a versatile set of transcriptional regulators
Abstract
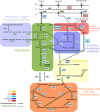
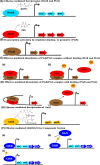
Bacteria of the genus Pseudomonas are widespread in nature. In the last decades, members of this genus, especially Pseudomonas aeruginosa and Pseudomonas putida, have acquired great interest because of their interactions with higher organisms. Pseudomonas aeruginosa is an opportunistic pathogen that colonizes the lung of cystic fibrosis patients, while P. putida is a soil bacterium able to establish a positive interaction with the plant rhizosphere. Members of Pseudomonas genus have a robust metabolism for amino acids and organic acids as well as aromatic compounds; however, these microbes metabolize a very limited number of sugars. Interestingly, they have three-pronged metabolic system to generate 6-phosphogluconate from glucose suggesting an adaptation to efficiently consume this sugar. This review focuses on the description of the regulatory network of glucose utilization in Pseudomonas, highlighting the differences between P. putida and P. aeruginosa. Most interestingly, It is highlighted a functional link between glucose assimilation and exotoxin A production in P. aeruginosa. The physiological relevance of this connection remains unclear, and it needs to be established whether a similar relationship is also found in other bacteria.
© 2018 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Figures




Similar articles
-
Transcriptional Modulation of Transport- and Metabolism-Associated Gene Clusters Leading to Utilization of Benzoate in Preference to Glucose in Pseudomonas putida CSV86.Appl Environ Microbiol. 2017 Sep 15;83(19):e01280-17. doi: 10.1128/AEM.01280-17. Print 2017 Oct 1. Appl Environ Microbiol. 2017. PMID: 28733285 Free PMC article.
-
Hierarchical management of carbon sources is regulated similarly by the CbrA/B systems in Pseudomonas aeruginosa and Pseudomonas putida.Microbiology (Reading). 2014 Oct;160(Pt 10):2243-2252. doi: 10.1099/mic.0.078873-0. Epub 2014 Jul 16. Microbiology (Reading). 2014. PMID: 25031426
-
GC-MS-based 13C metabolic flux analysis resolves the parallel and cyclic glucose metabolism of Pseudomonas putida KT2440 and Pseudomonas aeruginosa PAO1.Metab Eng. 2019 Jul;54:35-53. doi: 10.1016/j.ymben.2019.01.008. Epub 2019 Mar 1. Metab Eng. 2019. PMID: 30831266
-
Becoming settlers: Elements and mechanisms for surface colonization by Pseudomonas putida.Environ Microbiol. 2023 Sep;25(9):1575-1593. doi: 10.1111/1462-2920.16385. Epub 2023 Apr 12. Environ Microbiol. 2023. PMID: 37045787 Review.
-
Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans-metabolism.Metab Eng. 2018 Nov;50:142-155. doi: 10.1016/j.ymben.2018.05.005. Epub 2018 May 16. Metab Eng. 2018. PMID: 29758287 Review.
Cited by
-
Insights into the susceptibility of Pseudomonas putida to industrially relevant aromatic hydrocarbons that it can synthesize from sugars.Microb Cell Fact. 2023 Feb 2;22(1):22. doi: 10.1186/s12934-023-02028-y. Microb Cell Fact. 2023. PMID: 36732770 Free PMC article.
-
A multilocus sequence typing scheme of Pseudomonas putida for clinical and environmental isolates.Sci Rep. 2019 Sep 27;9(1):13980. doi: 10.1038/s41598-019-50299-6. Sci Rep. 2019. PMID: 31562354 Free PMC article.
-
Full Transcriptomic Response of Pseudomonas aeruginosa to an Inulin-Derived Fructooligosaccharide.Front Microbiol. 2020 Feb 20;11:202. doi: 10.3389/fmicb.2020.00202. eCollection 2020. Front Microbiol. 2020. PMID: 32153524 Free PMC article.
-
Engineered Passive Glucose Uptake in Pseudomonas taiwanensis VLB120 Increases Resource Efficiency for Bioproduction.Microb Biotechnol. 2025 Jan;18(1):e70095. doi: 10.1111/1751-7915.70095. Microb Biotechnol. 2025. PMID: 39871105 Free PMC article.
-
Gene expression reprogramming of Pseudomonas alloputida in response to arginine through the transcriptional regulator ArgR.Microbiology (Reading). 2024 Mar;170(3):001449. doi: 10.1099/mic.0.001449. Microbiology (Reading). 2024. PMID: 38511653 Free PMC article.
References
-
- Bateman, A. (1999) The SIS domain: a phosphosugar‐binding domain. Trends Biochem Sci 24: 94–95. - PubMed
-
- Braga, R. , Hecquet, L. , and Blonski, C. (2004) Slow‐binding inhibition of 2‐keto‐3 deoxy‐6phosphogluconate (KDPG) aldolase. Bioorg Med Chem 12: 2965–2972. - PubMed
-
- Colmer, J.A. , and Hamood, A.N. (1998) Characterization of ptxS, a Pseudomonas aeruginosa gene which interferes with the effect of the exotoxin A positive regulatory gene, ptxR . Mol Gen Genet 258: 250–259. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

