Proteomics and transcriptomics analyses of ataxia telangiectasia cells treated with Dexamethasone
- PMID: 29608596
- PMCID: PMC5880408
- DOI: 10.1371/journal.pone.0195388
Proteomics and transcriptomics analyses of ataxia telangiectasia cells treated with Dexamethasone
Abstract
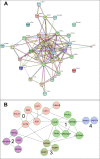
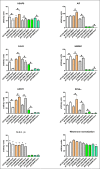
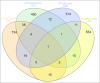
Ataxia telangiectasia (A-T) is an incurable and rare hereditary syndrome. In recent times, treatment with glucocorticoid analogues has been shown to improve the neurological symptoms that characterize this condition, but the molecular mechanism of action of these analogues remains unknown. Hence, the aim of this study was to gain insight into the molecular mechanism of action of glucocorticoid analogues in the treatment of A-T by investigating the role of Dexamethasone (Dexa) in A-T lymphoblastoid cell lines. We used 2DE and tandem MS to identify proteins that were influenced by the drug in A-T cells but not in healthy cells. Thirty-four proteins were defined out of a total of 746±63. Transcriptome analysis was performed by microarray and showed the differential expression of 599 A-T and 362 wild type (WT) genes and a healthy un-matching between protein abundance and the corresponding gene expression variation. The proteomic and transcriptomic profiles allowed the network pathway analysis to pinpoint the biological and molecular functions affected by Dexamethasone in Dexa-treated cells. The present integrated study provides evidence of the molecular mechanism of action of Dexamethasone in an A-T cellular model but also the broader effects of the drug in other tested cell lines.
Conflict of interest statement
Figures




References
-
- Savitsky K, Sfez S, Tagle DA, Ziv Y, Sartiel A, Collins FS, et al. (1995) The complete sequence of the coding region of the ATM gene reveals similarity to cell cycle regulators in different species. HumMolGenet 4: 2025–2032. - PubMed
-
- Abraham RT (2004) PI 3-kinase related kinases: 'big' players in stress-induced signaling pathways. DNA Repair (Amst) 3: 883–887. - PubMed
-
- Shiloh Y, Ziv Y (2013) The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. NatRevMolCell Biol 14: 197–210. - PubMed
-
- Chaudhary MW, Al-Baradie RS (2014) Ataxia-telangiectasia: future prospects. Appl Clin Genet 7: 159–167. doi: 10.2147/TACG.S35759 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

