The Prevalence and Evolutionary Conservation of Inverted Repeats in Proteobacteria
- PMID: 29608719
- PMCID: PMC5941160
- DOI: 10.1093/gbe/evy044
The Prevalence and Evolutionary Conservation of Inverted Repeats in Proteobacteria
Abstract
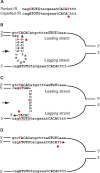
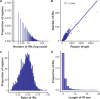
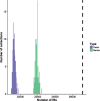
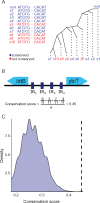
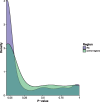
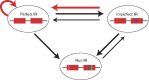
Perfect short inverted repeats (IRs) are known to be enriched in a variety of bacterial and eukaryotic genomes. Currently, it is unclear whether perfect IRs are conserved over evolutionary time scales. In this study, we aimed to characterize the prevalence and evolutionary conservation of IRs across 20 proteobacterial strains. We first identified IRs in Escherichia coli K-12 substr MG1655 and showed that they are overabundant. We next aimed to test whether this overabundance is reflected in the conservation of IRs over evolutionary time scales. To this end, for each perfect IR identified in E. coli MG1655, we collected orthologous sequences from related proteobacterial genomes. We next quantified the evolutionary conservation of these IRs, that is, the presence of the exact same IR across orthologous regions. We observed high conservation of perfect IRs: out of the 234 examined orthologous regions, 145 were more conserved than expected, which is statistically significant even after correcting for multiple testing. Our results together with previous experimental findings support a model in which imperfect IRs are corrected to perfect IRs in a preferential manner via a template switching mechanism.
Figures
Similar articles
-
Requirement or exclusion of inverted repeat sequences with cruciform-forming potential in Escherichia coli revealed by genome-wide analyses.Curr Genet. 2018 Aug;64(4):945-958. doi: 10.1007/s00294-018-0815-y. Epub 2018 Feb 27. Curr Genet. 2018. PMID: 29484452 Free PMC article.
-
A strong structural correlation between short inverted repeat sequences and the polyadenylation signal in yeast and nucleosome exclusion by these inverted repeats.Curr Genet. 2019 Apr;65(2):575-590. doi: 10.1007/s00294-018-0907-8. Epub 2018 Nov 29. Curr Genet. 2019. PMID: 30498953 Free PMC article.
-
Complex analyses of inverted repeats in mitochondrial genomes revealed their importance and variability.Bioinformatics. 2018 Apr 1;34(7):1081-1085. doi: 10.1093/bioinformatics/btx729. Bioinformatics. 2018. PMID: 29126205 Free PMC article.
-
Core-genome scaffold comparison reveals the prevalence that inversion events are associated with pairs of inverted repeats.BMC Genomics. 2017 Mar 29;18(1):268. doi: 10.1186/s12864-017-3655-0. BMC Genomics. 2017. PMID: 28356070 Free PMC article.
-
DNA repeat sequences: diversity and versatility of functions.Curr Genet. 2017 Jun;63(3):411-416. doi: 10.1007/s00294-016-0654-7. Epub 2016 Oct 14. Curr Genet. 2017. PMID: 27743028 Review.
Cited by
-
Protein innovation through template switching in the Saccharomyces cerevisiae lineage.Sci Rep. 2021 Nov 19;11(1):22558. doi: 10.1038/s41598-021-01736-y. Sci Rep. 2021. PMID: 34799587 Free PMC article.
-
Challenging the Importance of Plastid Genome Structure Conservation: New Insights From Euglenophytes.Mol Biol Evol. 2022 Dec 5;39(12):msac255. doi: 10.1093/molbev/msac255. Mol Biol Evol. 2022. PMID: 36403966 Free PMC article.
-
Generation of de novo miRNAs from template switching during DNA replication.Proc Natl Acad Sci U S A. 2023 Dec 5;120(49):e2310752120. doi: 10.1073/pnas.2310752120. Epub 2023 Nov 29. Proc Natl Acad Sci U S A. 2023. PMID: 38019864 Free PMC article.
-
Interaction of Proteins with Inverted Repeats and Cruciform Structures in Nucleic Acids.Int J Mol Sci. 2022 May 31;23(11):6171. doi: 10.3390/ijms23116171. Int J Mol Sci. 2022. PMID: 35682854 Free PMC article. Review.
-
PCIR: a database of Plant Chloroplast Inverted Repeats.Database (Oxford). 2019 Jan 1;2019:baz127. doi: 10.1093/database/baz127. Database (Oxford). 2019. PMID: 31696928 Free PMC article.
References
-
- Aris-Brosou S, Rodrigue N, Anisimova M.. 2012. The essentials of computational molecular evolution. Methods Mol Biol. 855:111–152. - PubMed
-
- Bissler JJ. 1998. DNA inverted repeats and human disease. Front Biosci. 3(4):d408–d418. - PubMed
-
- Blattner FR,, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277(5331):1453–1462. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

