Comparative Serum Challenges Show Divergent Patterns of Gene Expression and Open Chromatin in Human and Chimpanzee
- PMID: 29608722
- PMCID: PMC5848805
- DOI: 10.1093/gbe/evy041
Comparative Serum Challenges Show Divergent Patterns of Gene Expression and Open Chromatin in Human and Chimpanzee
Abstract
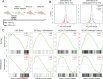
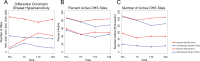
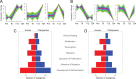
Humans experience higher rates of age-associated diseases than our closest living evolutionary relatives, chimpanzees. Environmental factors can explain many of these increases in disease risk, but species-specific genetic changes can also play a role. Alleles that confer increased disease susceptibility later in life can persist in a population in the absence of selective pressure if those changes confer positive adaptation early in life. One age-associated disease that disproportionately affects humans compared with chimpanzees is epithelial cancer. Here, we explored genetic differences between humans and chimpanzees in a well-defined experimental assay that mimics gene expression changes that happen during cancer progression: A fibroblast serum challenge. We used this assay with fibroblasts isolated from humans and chimpanzees to explore species-specific differences in gene expression and chromatin state with RNA-Seq and DNase-Seq. Our data reveal that human fibroblasts increase expression of genes associated with wound healing and cancer pathways; in contrast, chimpanzee gene expression changes are not concentrated around particular functional categories. Chromatin accessibility dramatically increases in human fibroblasts, yet decreases in chimpanzee cells during the serum response. Many regions of opening and closing chromatin are in close proximity to genes encoding transcription factors or genes involved in wound healing processes, further supporting the link between changes in activity of regulatory elements and changes in gene expression. Together, these expression and open chromatin data show that humans and chimpanzees have dramatically different responses to the same physiological stressor, and how a core physiological process can evolve quickly over relatively short evolutionary time scales.
Figures

References
-
- Abate-Shen C. 2002. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2(10):777–785. - PubMed
-
- American Cancer Society. 2016. Cancer Facts & Figures 2016. Atlanta, GA: American Cancer Society; 2016.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

