Higher Rates of Protein Evolution in the Self-Fertilizing Plant Arabidopsis thaliana than in the Out-Crossers Arabidopsis lyrata and Arabidopsis halleri
- PMID: 29608724
- PMCID: PMC5865523
- DOI: 10.1093/gbe/evy053
Higher Rates of Protein Evolution in the Self-Fertilizing Plant Arabidopsis thaliana than in the Out-Crossers Arabidopsis lyrata and Arabidopsis halleri
Abstract
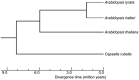
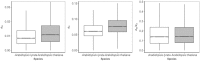
The common transition from out-crossing to self-fertilization in plants decreases effective population size. This is expected to result in a reduced efficacy of natural selection and in increased rates of protein evolution in selfing plants compared with their outcrossing congeners. Prior analyses, based on a very limited number of genes, detected no differences between the rates of protein evolution in the selfing Arabidopsis thaliana compared with the out-crosser Arabidopsis lyrata. Here, we reevaluate this trend using the complete genomes of A. thaliana, A. lyrata, Arabidopsis halleri, and the outgroups Capsella rubella and Thellungiella parvula. Our analyses indicate slightly but measurably higher nonsynonymous divergences (dN), synonymous divergences (dS) and dN/dS ratios in A. thaliana compared with the other Arabidopsis species, indicating that purifying selection is indeed less efficacious in A. thaliana.
Figures
Similar articles
-
Rates and patterns of molecular evolution in inbred and outbred Arabidopsis.Mol Biol Evol. 2002 Sep;19(9):1407-20. doi: 10.1093/oxfordjournals.molbev.a004204. Mol Biol Evol. 2002. PMID: 12200469
-
Reduced efficacy of natural selection on codon usage bias in selfing Arabidopsis and Capsella species.Genome Biol Evol. 2011;3:868-80. doi: 10.1093/gbe/evr085. Epub 2011 Aug 18. Genome Biol Evol. 2011. PMID: 21856647 Free PMC article.
-
The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr.Mol Biol Evol. 2006 Sep;23(9):1741-50. doi: 10.1093/molbev/msl042. Epub 2006 Jun 16. Mol Biol Evol. 2006. PMID: 16782760
-
Genomic studies of adaptive evolution in outcrossing Arabidopsis species.Curr Opin Plant Biol. 2017 Apr;36:9-14. doi: 10.1016/j.pbi.2016.11.018. Epub 2016 Dec 16. Curr Opin Plant Biol. 2017. PMID: 27988391 Review.
-
Beyond the thale: comparative genomics and genetics of Arabidopsis relatives.Nat Rev Genet. 2015 May;16(5):285-98. doi: 10.1038/nrg3883. Epub 2015 Apr 9. Nat Rev Genet. 2015. PMID: 25854181 Review.
Cited by
-
Paternally Expressed Imprinted Genes under Positive Darwinian Selection in Arabidopsis thaliana.Mol Biol Evol. 2019 Jun 1;36(6):1239-1253. doi: 10.1093/molbev/msz063. Mol Biol Evol. 2019. PMID: 30913563 Free PMC article.
-
Fixation of Expression Divergences by Natural Selection in Arabidopsis Coding Genes.Int J Mol Sci. 2024 Dec 22;25(24):13710. doi: 10.3390/ijms252413710. Int J Mol Sci. 2024. PMID: 39769472 Free PMC article.
-
Weaker selection on genes with treatment-specific expression consistent with a limit on plasticity evolution in Arabidopsis thaliana.Genetics. 2023 May 26;224(2):iyad074. doi: 10.1093/genetics/iyad074. Genetics. 2023. PMID: 37094602 Free PMC article.
-
Stop and go signals at the stigma-pollen interface of the Brassicaceae.Plant Physiol. 2023 Sep 22;193(2):927-948. doi: 10.1093/plphys/kiad301. Plant Physiol. 2023. PMID: 37423711 Free PMC article. No abstract available.
-
Comparative analyses of the Hymenoscyphus fraxineus and Hymenoscyphus albidus genomes reveals potentially adaptive differences in secondary metabolite and transposable element repertoires.BMC Genomics. 2021 Jul 4;22(1):503. doi: 10.1186/s12864-021-07837-2. BMC Genomics. 2021. PMID: 34217229 Free PMC article.
References
-
- Alvarez-Ponce D. 2014. Why proteins evolve at different rates: the determinants of proteins’ rates of evolution In: Fares MA, editor. Natural selection: methods and applications. London: CRC press; p. 126–178.
-
- Bechsgaard JS, Castric V, Charlesworth D, Vekemans X, Schierup MH.. 2006. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol Biol Evol. 23(9):1741–1750. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

