NADPH-dependent and -independent disulfide reductase systems
- PMID: 29609022
- PMCID: PMC6165701
- DOI: 10.1016/j.freeradbiomed.2018.03.051
NADPH-dependent and -independent disulfide reductase systems
Abstract
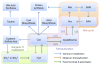
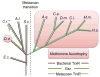
Over the past seven decades, research on autotrophic and heterotrophic model organisms has defined how the flow of electrons ("reducing power") from high-energy inorganic sources, through biological systems, to low-energy inorganic products like water, powers all of Life's processes. Universally, an initial major biological recipient of these electrons is nicotinamide adenine dinucleotide-phosphate, which thereby transits from an oxidized state (NADP+) to a reduced state (NADPH). A portion of this reducing power is then distributed via the cellular NADPH-dependent disulfide reductase systems as sequential reductions of disulfide bonds. Along the disulfide reduction pathways, some enzymes have active sites that use the selenium-containing amino acid, selenocysteine, in place of the common but less reactive sulfur-containing cysteine. In particular, the mammalian/metazoan thioredoxin systems are usually selenium-dependent as, across metazoan phyla, most thioredoxin reductases are selenoproteins. Among the roles of the NADPH-dependent disulfide reductase systems, the most universal is that they provide the reducing power for the production of DNA precursors by ribonucleotide reductase (RNR). Some studies, however, have uncovered examples of NADPH-independent disulfide reductase systems that can also support RNR. These systems are summarized here and their implications are discussed.
Keywords: Cysteine; Glutathione reductase; Methionine cycle; NADPH; Ribonucleotide reductase; Sulfur amino acid; Thioredoxin reductase; Transsulfuration.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures




References
-
- Buchanan BB. Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system. Perspective on its discovery, present status, and future development. Arch Biochem Biophys. 1991;288(1):1–9. - PubMed
-
- Calvin M, Benson AA. The Path of Carbon in Photosynthesis. Science. 1948;107(2784):476–80. - PubMed
-
- Bassham JA, Benson AA, Kay LD, Harris AZ, Wilson AT, Calvin M. The Path of Carbon in Photosynthesis. XXI. The Cyclic Regeneration of Carbon Dioxide Acceptor1. Journal of the American Chemical Society. 1954;76(7):1760–1770.
-
- Margittai É, Bánhegyi G. Isocitrate dehydrogenase: A NADPH-generating enzyme in the lumen of the endoplasmic reticulum. Archives of Biochemistry and Biophysics. 2008;471(2):184–190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

