Mycobacterium tuberculosis Transfer RNA Induces IL-12p70 via Synergistic Activation of Pattern Recognition Receptors within a Cell Network
- PMID: 29610140
- PMCID: PMC5916334
- DOI: 10.4049/jimmunol.1701733
Mycobacterium tuberculosis Transfer RNA Induces IL-12p70 via Synergistic Activation of Pattern Recognition Receptors within a Cell Network
Abstract
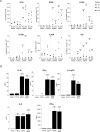
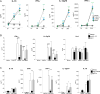
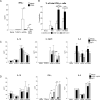
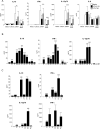
Upon recognition of a microbial pathogen, the innate and adaptive immune systems are linked to generate a cell-mediated immune response against the foreign invader. The culture filtrate of Mycobacterium tuberculosis contains ligands, such as M. tuberculosis tRNA, that activate the innate immune response and secreted Ags recognized by T cells to drive adaptive immune responses. In this study, bioinformatics analysis of gene-expression profiles derived from human PBMCs treated with distinct microbial ligands identified a mycobacterial tRNA-induced innate immune network resulting in the robust production of IL-12p70, a cytokine required to instruct an adaptive Th1 response for host defense against intracellular bacteria. As validated by functional studies, this pathway contained a feed-forward loop, whereby the early production of IL-18, type I IFNs, and IL-12p70 primed NK cells to respond to IL-18 and produce IFN-γ, enhancing further production of IL-12p70. Mechanistically, tRNA activates TLR3 and TLR8, and this synergistic induction of IL-12p70 was recapitulated by the addition of a specific TLR8 agonist with a TLR3 ligand to PBMCs. These data indicate that M. tuberculosis tRNA activates a gene network involving the integration of multiple innate signals, including types I and II IFNs, as well as distinct cell types to induce IL-12p70.
Copyright © 2018 by The American Association of Immunologists, Inc.
Figures




References
-
- Bloom BR, Murray CJL. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. - PubMed
-
- Seibert FB. The isolation of a crystalline protein with tuberculin activity. Science. 1926;63:619c–620. - PubMed
-
- Long ER, Seibert FB. The chemical composition of the active principle of tuberculin. VII. The evidence that the active principle is a protein. Am. Rev. Tuberc. 1926;13:448.
-
- Seibert FB. X. The isolation in crystalline form and identification of the active principle of tuberculin. Am Rev Tuberc. 1928;17:402–421.
-
- Seibert FB. The isolation and properties of the purified protein derivative of tuberculin. Am. Rev. Tuberc. 1934;30:713–720.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

