Spatial organization and dynamics of RNase E and ribosomes in Caulobacter crescentus
- PMID: 29610352
- PMCID: PMC5910860
- DOI: 10.1073/pnas.1721648115
Spatial organization and dynamics of RNase E and ribosomes in Caulobacter crescentus
Abstract
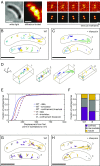
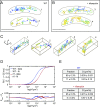
We report the dynamic spatial organization of Caulobacter crescentus RNase E (RNA degradosome) and ribosomal protein L1 (ribosome) using 3D single-particle tracking and superresolution microscopy. RNase E formed clusters along the central axis of the cell, while weak clusters of ribosomal protein L1 were deployed throughout the cytoplasm. These results contrast with RNase E and ribosome distribution in Escherichia coli, where RNase E colocalizes with the cytoplasmic membrane and ribosomes accumulate in polar nucleoid-free zones. For both RNase E and ribosomes in Caulobacter, we observed a decrease in confinement and clustering upon transcription inhibition and subsequent depletion of nascent RNA, suggesting that RNA substrate availability for processing, degradation, and translation facilitates confinement and clustering. Importantly, RNase E cluster positions correlated with the subcellular location of chromosomal loci of two highly transcribed rRNA genes, suggesting that RNase E's function in rRNA processing occurs at the site of rRNA synthesis. Thus, components of the RNA degradosome and ribosome assembly are spatiotemporally organized in Caulobacter, with chromosomal readout serving as the template for this organization.
Keywords: RNA degradation; RNA processing; ribosomes; single-molecule tracking; superresolution microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Mackie GA. RNase E: At the interface of bacterial RNA processing and decay. Nat Rev Microbiol. 2013;11:45–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

