Genomic profiling of dedifferentiated liposarcoma compared to matched well-differentiated liposarcoma reveals higher genomic complexity and a common origin
- PMID: 29610390
- PMCID: PMC5880260
- DOI: 10.1101/mcs.a002386
Genomic profiling of dedifferentiated liposarcoma compared to matched well-differentiated liposarcoma reveals higher genomic complexity and a common origin
Abstract
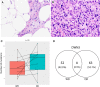
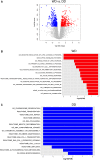
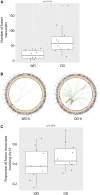
Well-differentiated (WD) liposarcoma is a low-grade mesenchymal tumor with features of mature adipocytes and high propensity for local recurrence. Often, WD patients present with or later progress to a higher-grade nonlipogenic form known as dedifferentiated (DD) liposarcoma. These DD tumors behave more aggressively and can metastasize. Both WD and DD liposarcomas harbor neochromosomes formed from amplifications and rearrangements of Chr 12q that encode oncogenes (MDM2, CDK4, and YEATS2) and adipocytic differentiation factors (HMGA2 and CPM) However, genomic changes associated with progression from WD to DD have not been well-defined. Therefore, we selected patients with matched WD and DD tumors for extensive genomic profiling in order to understand their clonal relationships and to delineate any defining alterations for each entity. Exome and transcriptomic sequencing was performed for 17 patients with both WD and DD diagnoses. Somatic point and copy-number alterations were integrated with transcriptional analyses to determine subtype-associated genomic features and pathways. The results were, on average, that only 8.3% of somatic mutations in WD liposarcoma were shared with their cognate DD component. DD tumors had higher numbers of somatic copy-number losses, amplifications involving Chr 12q, and fusion transcripts than WD tumors. HMGA2 and CPM rearrangements occur more frequently in DD components. The shared somatic mutations indicate a clonal origin for matched WD and DD tumors and show early divergence with ongoing genomic instability due to continual generation and selection of neochromosomes. Stochastic generation and subsequent expression of fusion transcripts from the neochromosome that involve adipogenesis genes such as HMGA2 and CPM may influence the differentiation state of the subsequent tumor.
Keywords: overgrowth.
© 2018 Beird et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
-
- Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, et al. 2013. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39: 782–795. - PubMed
-
- Coindre JM, Pédeutour F, Aurias A. 2010. Well-differentiated and dedifferentiated liposarcomas. Virchows Arch 456: 167–179. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
