Effect of Ca2+ on the promiscuous target-protein binding of calmodulin
- PMID: 29614072
- PMCID: PMC5898786
- DOI: 10.1371/journal.pcbi.1006072
Effect of Ca2+ on the promiscuous target-protein binding of calmodulin
Abstract
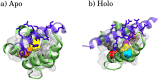
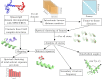
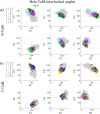
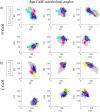
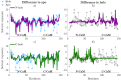
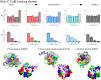
Calmodulin (CaM) is a calcium sensing protein that regulates the function of a large number of proteins, thus playing a crucial part in many cell signaling pathways. CaM has the ability to bind more than 300 different target peptides in a Ca2+-dependent manner, mainly through the exposure of hydrophobic residues. How CaM can bind a large number of targets while retaining some selectivity is a fascinating open question. Here, we explore the mechanism of CaM selective promiscuity for selected target proteins. Analyzing enhanced sampling molecular dynamics simulations of Ca2+-bound and Ca2+-free CaM via spectral clustering has allowed us to identify distinct conformational states, characterized by interhelical angles, secondary structure determinants and the solvent exposure of specific residues. We searched for indicators of conformational selection by mapping solvent exposure of residues in these conformational states to contacts in structures of CaM/target peptide complexes. We thereby identified CaM states involved in various binding classes arranged along a depth binding gradient. Binding Ca2+ modifies the accessible hydrophobic surface of the two lobes and allows for deeper binding. Apo CaM indeed shows shallow binding involving predominantly polar and charged residues. Furthermore, binding to the C-terminal lobe of CaM appears selective and involves specific conformational states that can facilitate deep binding to target proteins, while binding to the N-terminal lobe appears to happen through a more flexible mechanism. Thus the long-ranged electrostatic interactions of the charged residues of the N-terminal lobe of CaM may initiate binding, while the short-ranged interactions of hydrophobic residues in the C-terminal lobe of CaM may account for selectivity. This work furthers our understanding of the mechanism of CaM binding and selectivity to different target proteins and paves the way towards a comprehensive model of CaM selectivity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Stevens FC. Calmodulin: an introduction. Canadian Journal of Biochemistry and Cell Biology. 1983;61(8):906–910. doi: 10.1139/o83-115 - DOI - PubMed
-
- Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Solution structure of calcium-free calmodulin. Nature Structural & Molecular Biology. 1995;2(9):768–776. doi: 10.1038/nsb0995-768 - DOI - PubMed
-
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092 - DOI - PMC - PubMed
-
- Smith DMA, Straatsma TP, Squier TC. Retention of Conformational Entropy upon Calmodulin Binding to Target Peptides Is Driven by Transient Salt Bridges. Biophysical Journal. 2012;103(7):1576–1584. doi: 10.1016/j.bpj.2012.08.037 - DOI - PMC - PubMed
-
- Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 1997;11(5):331–340. doi: 10.1096/fasebj.11.5.9141499 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

