Defining essential elements and genetic interactions of the yeast Lsm2-8 ring and demonstration that essentiality of Lsm2-8 is bypassed via overexpression of U6 snRNA or the U6 snRNP subunit Prp24
- PMID: 29615482
- PMCID: PMC5959253
- DOI: 10.1261/rna.066175.118
Defining essential elements and genetic interactions of the yeast Lsm2-8 ring and demonstration that essentiality of Lsm2-8 is bypassed via overexpression of U6 snRNA or the U6 snRNP subunit Prp24
Abstract
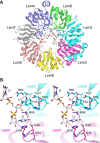
A seven-subunit Lsm2-8 protein ring assembles on the U-rich 3' end of the U6 snRNA. A structure-guided mutational analysis of the Saccharomyces cerevisiae Lsm2-8 ring affords new insights to structure-function relations and genetic interactions of the Lsm subunits. Alanine scanning of 39 amino acids comprising the RNA-binding sites or intersubunit interfaces of Lsm2, Lsm3, Lsm4, Lsm5, and Lsm8 identified only one instance of lethality (Lsm3-R69A) and one severe growth defect (Lsm2-R63A), both involving amino acids that bind the 3'-terminal UUU trinucleotide. All other Ala mutations were benign with respect to vegetative growth. Tests of 235 pairwise combinations of benign Lsm mutants identified six instances of inter-Lsm synthetic lethality and 45 cases of nonlethal synthetic growth defects. Thus, Lsm2-8 ring function is buffered by a network of internal genetic redundancies. A salient finding was that otherwise lethal single-gene deletions lsm2Δ, lsm3Δ, lsm4Δ, lsm5, and lsm8Δ were rescued by overexpression of U6 snRNA from a high-copy plasmid. Moreover, U6 overexpression rescued myriad lsmΔ lsmΔ double-deletions and lsmΔ lsmΔ lsmΔ triple-deletions. We find that U6 overexpression also rescues a lethal deletion of the U6 snRNP protein subunit Prp24 and that Prp24 overexpression bypasses the essentiality of the U6-associated Lsm subunits. Our results indicate that abetting U6 snRNA is the only essential function of the yeast Lsm2-8 proteins.
Keywords: Lsm proteins; Prp24; U6 snRNA; mRNA splicing.
© 2018 Roth et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures







References
-
- Chan SP, Kao DI, Tsai WY, Cheng SC. 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
