Yeast Gup1(2) Proteins Are Homologues of the Hedgehog Morphogens Acyltransferases HHAT(L): Facts and Implications
- PMID: 29615596
- PMCID: PMC5831804
- DOI: 10.3390/jdb4040033
Yeast Gup1(2) Proteins Are Homologues of the Hedgehog Morphogens Acyltransferases HHAT(L): Facts and Implications
Abstract
In multiple tissues, the Hedgehog secreted morphogen activates in the receiving cells a pathway involved in cell fate, proliferation and differentiation in the receiving cells. This pathway is particularly important during embryogenesis. The protein HHAT (Hedgehog O-acyltransferase) modifies Hh morphogens prior to their secretion, while HHATL (Hh O-acyltransferase-like) negatively regulates the pathway. HHAT and HHATL are homologous to Saccharomyces cerevisiae Gup2 and Gup1, respectively. In yeast, Gup1 is associated with a high number and diversity of biological functions, namely polarity establishment, secretory/endocytic pathway functionality, vacuole morphology and wall and membrane composition, structure and maintenance. Phenotypes underlying death, morphogenesis and differentiation are also included. Paracrine signalling, like the one promoted by the Hh pathway, has not been shown to occur in microbial communities, despite the fact that large aggregates of cells like biofilms or colonies behave as proto-tissues. Instead, these have been suggested to sense the population density through the secretion of quorum-sensing chemicals. This review focuses on Gup1/HHATL and Gup2/HHAT proteins. We review the functions and physiology associated with these proteins in yeasts and higher eukaryotes. We suggest standardisation of the presently chaotic Gup-related nomenclature, which includes KIAA117, c3orf3, RASP, Skinny, Sightless and Central Missing, in order to avoid the disclosure of otherwise unnoticed information.
Keywords: GUP; HHAT; Hedgehog; morphogenesis; yeast.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
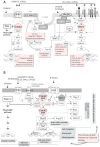
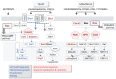

 ), glucosamine (✩) and ethanolamine-P (☐). Dark thick scrawls represent peptide chains. PI: phosphatidylinositol; IPC: inositol phosphoceramide. Types B, C and D were defined from bands obtained using TLC [136].
), glucosamine (✩) and ethanolamine-P (☐). Dark thick scrawls represent peptide chains. PI: phosphatidylinositol; IPC: inositol phosphoceramide. Types B, C and D were defined from bands obtained using TLC [136].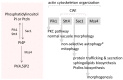

References
-
- Verduyckt M., Vignaud H., Bynens T., van den Brande J., Franssens V., Cullin C., Winderickx J. Yeast as a model for Alzheimer’s disease: Latest studies and advanced strategies. Meth. Mol. Biol. 2016;1303:197–215. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

