Epigenetic alterations are associated with monocyte immune dysfunctions in HIV-1 infection
- PMID: 29615725
- PMCID: PMC5882962
- DOI: 10.1038/s41598-018-23841-1
Epigenetic alterations are associated with monocyte immune dysfunctions in HIV-1 infection
Abstract
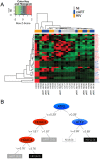
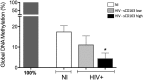
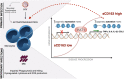
Monocytes are key cells in the immune dysregulation observed during human immunodeficiency virus (HIV) infection. The events that take place specifically in monocytes may contribute to the systemic immune dysfunction characterized by excessive immune activation in infected individuals, which directly correlates with pathogenesis and progression of the disease. Here, we investigated the immune dysfunction in monocytes from untreated and treated HIV + patients and associated these findings with epigenetic changes. Monocytes from HIV patients showed dysfunctional ability of phagocytosis and killing, and exhibited dysregulated cytokines and reactive oxygen species production after M. tuberculosis challenge in vitro. In addition, we showed that the expression of enzymes responsible for epigenetic changes was altered during HIV infection and was more prominent in patients that had high levels of soluble CD163 (sCD163), a newly identified plasmatic HIV progression biomarker. Among the enzymes, histone acetyltransferase 1 (HAT1) was the best epigenetic biomarker correlated with HIV - sCD163 high patients. In conclusion, we confirmed that HIV impairs effector functions of monocytes and these alterations are associated with epigenetic changes that once identified could be used as targets in therapies aiming the reduction of the systemic activation state found in HIV patients.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Espíndola, M. S. et al. HIV infection: focus on the innate immune cells. Immunologic Research, 1–15 10.1007/s12026-016-8862-2 (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

