Centrifugation-free extraction of circulating nucleic acids using immiscible liquid under vacuum pressure
- PMID: 29615736
- PMCID: PMC5883035
- DOI: 10.1038/s41598-018-23766-9
Centrifugation-free extraction of circulating nucleic acids using immiscible liquid under vacuum pressure
Abstract
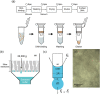
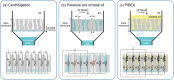
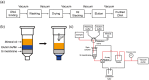
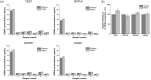
Extraction of cell-free DNA (cfDNA), which exists at an extremely low concentration in plasma, is a critical process for either targeted-sensing or massive sequencing of DNAs. However, such small amount of DNA cannot be fully obtained without high-speed centrifugation (<20,000 g). Here, we developed a centrifugation-free cfDNA extraction method and system that utilizes an immiscible solvent under single low vacuum pressure throughout the entire process. It has been named Pressure and Immiscibility-Based EXtraction (PIBEX). The amounts of extracted cfDNA by PIBEX were compared with those extracted by the conventional gold standards such as QIAGEN using quantitative PCR (qPCR). The PIBEX system showed equal performance regarding extraction amount and efficiency compared to the existing method. Because the PIBEX eliminates the troublous and repetitive centrifugation processes in DNA extraction, it can be further utilized in microfluidic-sample preparation systems for circulating nucleic acids, which would lead to an integrated sample-to-answer system in liquid biopsies.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

