De novo draft assembly of the Botrylloides leachii genome provides further insight into tunicate evolution
- PMID: 29615780
- PMCID: PMC5882950
- DOI: 10.1038/s41598-018-23749-w
De novo draft assembly of the Botrylloides leachii genome provides further insight into tunicate evolution
Abstract
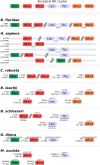
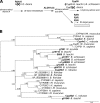
Tunicates are marine invertebrates that compose the closest phylogenetic group to the vertebrates. These chordates present a particularly diverse range of regenerative abilities and life-history strategies. Consequently, tunicates provide an extraordinary perspective into the emergence and diversity of these traits. Here we describe the genome sequencing, annotation and analysis of the Stolidobranchian Botrylloides leachii. We have produced a high-quality 159 Mb assembly, 82% of the predicted 194 Mb genome. Analysing genome size, gene number, repetitive elements, orthologs clustering and gene ontology terms show that B. leachii has a genomic architecture similar to that of most solitary tunicates, while other recently sequenced colonial ascidians have undergone genome expansion. In addition, ortholog clustering has identified groups of candidate genes for the study of colonialism and whole-body regeneration. By analysing the structure and composition of conserved gene linkages, we observed examples of cluster breaks and gene dispersions, suggesting that several lineage-specific genome rearrangements occurred during tunicate evolution. We also found lineage-specific gene gain and loss within conserved cell-signalling pathways. Such examples of genetic changes within conserved cell-signalling pathways commonly associated with regeneration and development that may underlie some of the diverse regenerative abilities observed in tunicates. Overall, these results provide a novel resource for the study of tunicates and of colonial ascidians.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Millar RH. The biology of ascidians. Advanced Marine Biology. 1971;9:1–100. doi: 10.1016/S0065-2881(08)60341-7. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

