Evolutionary pruning of transfer learned deep convolutional neural network for breast cancer diagnosis in digital breast tomosynthesis
- PMID: 29616660
- PMCID: PMC5967610
- DOI: 10.1088/1361-6560/aabb5b
Evolutionary pruning of transfer learned deep convolutional neural network for breast cancer diagnosis in digital breast tomosynthesis
Abstract
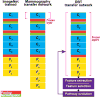
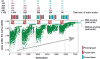
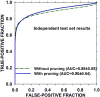
Deep learning models are highly parameterized, resulting in difficulty in inference and transfer learning for image recognition tasks. In this work, we propose a layered pathway evolution method to compress a deep convolutional neural network (DCNN) for classification of masses in digital breast tomosynthesis (DBT). The objective is to prune the number of tunable parameters while preserving the classification accuracy. In the first stage transfer learning, 19 632 augmented regions-of-interest (ROIs) from 2454 mass lesions on mammograms were used to train a pre-trained DCNN on ImageNet. In the second stage transfer learning, the DCNN was used as a feature extractor followed by feature selection and random forest classification. The pathway evolution was performed using genetic algorithm in an iterative approach with tournament selection driven by count-preserving crossover and mutation. The second stage was trained with 9120 DBT ROIs from 228 mass lesions using leave-one-case-out cross-validation. The DCNN was reduced by 87% in the number of neurons, 34% in the number of parameters, and 95% in the number of multiply-and-add operations required in the convolutional layers. The test AUC on 89 mass lesions from 94 independent DBT cases before and after pruning were 0.88 and 0.90, respectively, and the difference was not statistically significant (p > 0.05). The proposed DCNN compression approach can reduce the number of required operations by 95% while maintaining the classification performance. The approach can be extended to other deep neural networks and imaging tasks where transfer learning is appropriate.
Figures

References
-
- US FOOD & DRUG ADMINISTRATION: MQSA National Statistics. 2017 Link: www.fda.gov/Radiation-EmittingProducts/MammographyQualityStandardsActand... (accessed on: 07/03/2017)
-
- Anwar S, Hwang K, Sung W. Structured pruning of deep convolutional neural networks. ACM Journal on Emerging Technologies in Computing Systems (JETC) 2017;13:32.
-
- Chan H-P, Lo SC, Helvie M, Goodsitt MM, Cheng SNC, Adler DD. Recognition of mammographic microcalcifications with artificial neural network. Radiology. 1993;189(P):318.
-
- Chan H-P, Lo SCB, Sahiner B, Lam KL, Helvie MA. Computer-aided detection of mammographic microcalcifications: Pattern recognition with an artificial neural network. Medical Physics. 1995;22:1555–67. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
