Exosome-Mediated Transfer of Cancer Cell Resistance to Antiestrogen Drugs
- PMID: 29617321
- PMCID: PMC6017149
- DOI: 10.3390/molecules23040829
Exosome-Mediated Transfer of Cancer Cell Resistance to Antiestrogen Drugs
Abstract
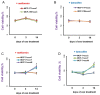
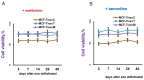
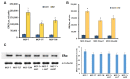
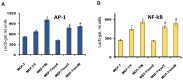
Exosomes are small vesicles which are produced by the cells and released into the surrounding space. They can transfer biomolecules into recipient cells. The main goal of the work was to study the exosome involvement in the cell transfer of hormonal resistance. The experiments were performed on in vitro cultured estrogen-dependent MCF-7 breast cancer cells and MCF-7 sublines resistant to SERM tamoxifen and/or biguanide metformin, which exerts its anti-proliferative effect, at least in a part, via the suppression of estrogen machinery. The exosomes were purified by differential ultracentrifugation, cell response to tamoxifen was determined by MTT test, and the level and activity of signaling proteins were determined by Western blot and reporter analysis. We found that the treatment of the parent MCF-7 cells with exosomes from the resistant cells within 14 days lead to the partial resistance of the MCF-7 cells to antiestrogen drugs. The primary resistant cells and the cells with the exosome-induced resistance were characterized with these common features: decrease in ERα activity and parallel activation of Akt and AP-1, NF-κB, and SNAIL1 transcriptional factors. In general, we evaluate the established results as the evidence of the possible exosome involvement in the transferring of the hormone/metformin resistance in breast cancer cells.
Keywords: SERM; breast cancer; exosomes; resistance; tamoxifen.
Conflict of interest statement
The authors declare no conflict of interest regarding this article.
Figures





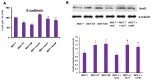
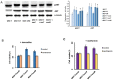
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

