Computational Analysis for the Rational Design of Anti-Amyloid Beta (Aβ) Antibodies
- PMID: 29617557
- PMCID: PMC5927393
- DOI: 10.1021/acs.jpcb.8b01837
Computational Analysis for the Rational Design of Anti-Amyloid Beta (Aβ) Antibodies
Abstract
Alzheimer's disease (AD) is a neurodegenerative disorder that lacks effective treatment options. Anti-amyloid beta (Aβ) antibodies are the leading drug candidates to treat AD, but the results of clinical trials have been disappointing. Introducing rational mutations into anti-Aβ antibodies to increase their effectiveness is a way forward, but the path to take is unclear. In this study, we demonstrate the use of computational fragment-based docking and MMPBSA binding free energy calculations in the analysis of anti-Aβ antibodies for rational drug design efforts. Our fragment-based docking method successfully predicts the emergence of the common EFRH epitope. MD simulations coupled with MMPBSA binding free energy calculations are used to analyze scenarios described in prior studies, and we computationally introduce rational mutations into PFA1 to predict mutations that can improve its binding affinity toward the pE3-Aβ3-8 form of Aβ. Two out of our four proposed mutations are predicted to stabilize binding. Our study demonstrates that a computational approach may lead to an improved drug candidate for AD in the future.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
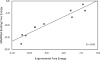





Similar articles
-
Active immunization with the peptide epitope vaccine Aβ3-10-KLH induces a Th2-polarized anti-Aβ antibody response and decreases amyloid plaques in APP/PS1 transgenic mice.Neurosci Lett. 2016 Nov 10;634:1-6. doi: 10.1016/j.neulet.2016.09.050. Epub 2016 Sep 28. Neurosci Lett. 2016. PMID: 27693663
-
Epitope structure and binding affinity of single chain llama anti-β-amyloid antibodies revealed by proteolytic excision affinity-mass spectrometry.J Mol Recognit. 2013 Jan;26(1):1-9. doi: 10.1002/jmr.2210. J Mol Recognit. 2013. PMID: 23280612
-
Deglycosylated anti-amyloid beta antibodies reduce microglial phagocytosis and cytokine production while retaining the capacity to induce amyloid beta sequestration.Eur J Neurosci. 2007 Nov;26(9):2458-68. doi: 10.1111/j.1460-9568.2007.05852.x. Epub 2007 Oct 23. Eur J Neurosci. 2007. PMID: 17970733
-
Immunotherapy for Alzheimer's disease.Acta Biochim Biophys Sin (Shanghai). 2012 Oct;44(10):807-14. doi: 10.1093/abbs/gms065. Epub 2012 Aug 16. Acta Biochim Biophys Sin (Shanghai). 2012. PMID: 22899646 Review.
-
Conformation-specific antibodies to target amyloid β oligomers and their application to immunotherapy for Alzheimer's disease.Biosci Biotechnol Biochem. 2014;78(8):1293-305. doi: 10.1080/09168451.2014.940275. Biosci Biotechnol Biochem. 2014. PMID: 25130729 Review.
Cited by
-
Recent Developments in Free Energy Calculations for Drug Discovery.Front Mol Biosci. 2021 Aug 11;8:712085. doi: 10.3389/fmolb.2021.712085. eCollection 2021. Front Mol Biosci. 2021. PMID: 34458321 Free PMC article. Review.
-
Robustness and Efficiency of Poisson-Boltzmann Modeling on Graphics Processing Units.J Chem Inf Model. 2019 Jan 28;59(1):409-420. doi: 10.1021/acs.jcim.8b00761. Epub 2018 Dec 31. J Chem Inf Model. 2019. PMID: 30550277 Free PMC article.
-
Molecular interactions between monoclonal oligomer-specific antibody 5E3 and its amyloid beta cognates.PLoS One. 2020 May 29;15(5):e0232266. doi: 10.1371/journal.pone.0232266. eCollection 2020. PLoS One. 2020. PMID: 32469918 Free PMC article.
References
-
- Iqbal K, del C Alonso A, Chen S, Chohan MO, El-Akkad E, Gong CX, Khatoon S, Li B, Liu F, Rahman A, et al. Tau pathology in Alzheimer disease and other tauopathies. Biochim Biophys Acta, Mol Basis Dis. 2005;1739(2–3):198–210. - PubMed
-
- Dobson CM. Getting out of shape. Nature. 2002;418:729–730. - PubMed
-
- Weiner HL, Frenkel D. Immunology and immunotherapy of Alzheimer’s disease. Nat Rev Immunol. 2006;6:404–416. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

