A survey and evaluation of Web-based tools/databases for variant analysis of TCGA data
- PMID: 29617727
- PMCID: PMC6781580
- DOI: 10.1093/bib/bby023
A survey and evaluation of Web-based tools/databases for variant analysis of TCGA data
Abstract
The Cancer Genome Atlas (TCGA) is a publicly funded project that aims to catalog and discover major cancer-causing genomic alterations with the goal of creating a comprehensive 'atlas' of cancer genomic profiles. The availability of this genome-wide information provides an unprecedented opportunity to expand our knowledge of tumourigenesis. Computational analytics and mining are frequently used as effective tools for exploring this byzantine series of biological and biomedical data. However, some of the more advanced computational tools are often difficult to understand or use, thereby limiting their application by scientists who do not have a strong computational background. Hence, it is of great importance to build user-friendly interfaces that allow both computational scientists and life scientists without a computational background to gain greater biological and medical insights. To that end, this survey was designed to systematically present available Web-based tools and facilitate the use TCGA data for cancer research.
Keywords: The Cancer Genome Atlas; bioinformatics tools; cancer; databases; survey.
© The Author(s) 2018. Published by Oxford University Press.
Figures

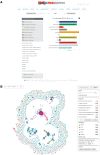



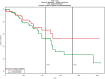
References
-
- Lander ES, Linton LM, Birren B, et al. . Initial sequencing and analysis of the human genome. Nature 2001;409(6822):860–921. - PubMed
-
- Venter JC, Adams MD, Myers EW, et al. . The sequence of the human genome. Science 2001;291:1304–51. - PubMed
-
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004;431:931–45. - PubMed
-
- Garraway LA. Genomics-Driven Oncology: framework for an Emerging Paradigm. J Clin Oncol 2013;31(15):1806–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

