Control of mRNA decapping by autoinhibition
- PMID: 29618050
- PMCID: PMC6158755
- DOI: 10.1093/nar/gky233
Control of mRNA decapping by autoinhibition
Abstract
5' mediated cytoplasmic RNA decay is a conserved cellular process in eukaryotes. While the functions of the structured core domains in this pathway are well-studied, the role of abundant intrinsically disordered regions (IDRs) is lacking. Here we reconstitute the Dcp1:Dcp2 complex containing a portion of the disordered C-terminus and show its activity is autoinhibited by linear interaction motifs. Enhancers of decapping (Edc) 1 and 3 cooperate to activate decapping by different mechanisms: Edc3 alleviates autoinhibition by binding IDRs and destabilizing an inactive form of the enzyme, whereas Edc1 stabilizes the transition state for catalysis. Both activators are required to fully stimulate an autoinhibited Dcp1:Dcp2 as Edc1 alone cannot overcome the decrease in activity attributed to the C-terminal extension. Our data provide a mechanistic framework for combinatorial control of decapping by protein cofactors, a principle that is likely conserved in multiple 5' mRNA decay pathways.
Figures

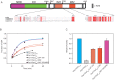

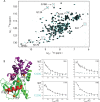
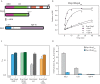
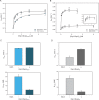


References
-
- Konarska M.M., Padgett R.A., Sharp P.A.. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984; 38:731–736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

