Review: Genomics of bull fertility
- PMID: 29618393
- PMCID: PMC6550321
- DOI: 10.1017/S1751731118000599
Review: Genomics of bull fertility
Abstract
Fertility is one of the most economically important traits in both beef and dairy cattle production; however, only female fertility is typically subjected to selection. Male and female fertility have only a small positive genetic correlation which is likely due to the existence of a relatively small number of genetic variants within each breed that cause embryonic and developmental losses. Genomic tools have been developed that allow the identification of lethal recessive loci based upon marker haplotypes. Selection against haplotypes harbouring lethal alleles in conjunction with selection to improve female fertility will result in an improvement in male fertility. Genomic selection has resulted in a two to fourfold increase in the rate of genetic improvement of most dairy traits in US Holstein cattle, including female fertility. Considering the rapidly increasing rate of adoption of high-throughput single nucleotide polymorphism genotyping in both the US dairy and beef industries, genomic selection should be the most effective of all currently available approaches to improve male fertility. However, male fertility phenotypes are not routinely recorded in natural service mating systems and when artificial insemination is used, semen doses may be titrated to lower post-thaw progressively motile sperm numbers for high-merit and high-demand bulls. Standardization of sperm dosages across bull studs for semen distributed from young bulls would allow the capture of sire conception rate phenotypes for young bulls that could be used to generate predictions of genetic merit for male fertility in both males and females. These data would allow genomic selection to be implemented for male fertility in addition to female fertility within the US dairy industry. While the rate of use of artificial insemination is much lower within the US beef industry, the adoption of sexed semen in the dairy industry has allowed dairy herds to select cows from which heifer replacements are produced and cows that are used to produce terminal crossbred bull calves sired by beef breed bulls. Capture of sire conception rate phenotypes in dairy herds utilizing sexed semen will contribute data enabling genomic selection for male fertility in beef cattle breeds. As the commercial sector of the beef industry increasingly adopts fixed-time artificial insemination, sire conception rate phenotypes can be captured to facilitate the development of estimates of genetic merit for male fertility within US beef breeds.
Keywords: bull fertility; candidate genes; genome-wide association study; genomic selection; quantitative trait loci.
Conflict of interest statement
Declaration of interest
The authors have no conflicts of interest.
Figures
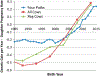
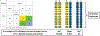

References
-
- Abdollahi-Arpanahi R, Morota G and Peñagaricano F 2017. Predicting bull fertility using genomic data and biological information. Journal of Dairy Science 100, 9656–9666. - PubMed
-
- Berry DP, Wall E and Pryce JE 2014. Genetics and genomics of reproductive performance in dairy and beef cattle. Animal 8 (suppl. 1), 105–121. - PubMed
-
- Card CJ, Krieger KE, Kaproth M and Sartini BL 2017. Oligo-dT selected spermatozoal transcript profiles differ among higher and lower fertility dairy sires. Animal Reproduction Science 177, 105–123. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

