Subtle Perturbations of the Maize Methylome Reveal Genes and Transposons Silenced by Chromomethylase or RNA-Directed DNA Methylation Pathways
- PMID: 29618467
- PMCID: PMC5982821
- DOI: 10.1534/g3.118.200284
Subtle Perturbations of the Maize Methylome Reveal Genes and Transposons Silenced by Chromomethylase or RNA-Directed DNA Methylation Pathways
Abstract
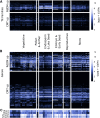
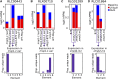
DNA methylation is a chromatin modification that can provide epigenetic regulation of gene and transposon expression. Plants utilize several pathways to establish and maintain DNA methylation in specific sequence contexts. The chromomethylase (CMT) genes maintain CHG (where H = A, C or T) methylation. The RNA-directed DNA methylation (RdDM) pathway is important for CHH methylation. Transcriptome analysis was performed in a collection of Zea mays lines carrying mutant alleles for CMT or RdDM-associated genes. While the majority of the transcriptome was not affected, we identified sets of genes and transposon families sensitive to context-specific decreases in DNA methylation in mutant lines. Many of the genes that are up-regulated in CMT mutant lines have high levels of CHG methylation, while genes that are differentially expressed in RdDM mutants are enriched for having nearby mCHH islands, implicating context-specific DNA methylation in the regulation of expression for a small number of genes. Many genes regulated by CMTs exhibit natural variation for DNA methylation and transcript abundance in a panel of diverse inbred lines. Transposon families with differential expression in the mutant genotypes show few defining features, though several families up-regulated in RdDM mutants show enriched expression in endosperm tissue, highlighting the potential importance for this pathway during reproduction. Taken together, our findings suggest that while the number of genes and transposon families whose expression is reproducibly affected by mild perturbations in context-specific methylation is small, there are distinct patterns for loci impacted by RdDM and CMT mutants.
Keywords: DNA methylation; RNA directed DNA methylation; chromomethylase; maize; transposable elements.
Copyright © 2018 Anderson et al.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
