Cyclase-associated protein 1 (CAP1) is a prenyl-binding partner of Rap1 GTPase
- PMID: 29618512
- PMCID: PMC5961064
- DOI: 10.1074/jbc.RA118.001779
Cyclase-associated protein 1 (CAP1) is a prenyl-binding partner of Rap1 GTPase
Erratum in
-
Correction: Cyclase-associated protein 1 (CAP1) is a prenyl-binding partner of Rap1 GTPase.J Biol Chem. 2018 Sep 7;293(36):13849. doi: 10.1074/jbc.AAC118.005290. J Biol Chem. 2018. PMID: 30194256 Free PMC article. No abstract available.
Abstract
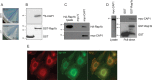
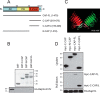
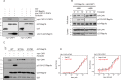
Rap1 proteins are members of the Ras subfamily of small GTPases involved in many biological responses, including adhesion, cell proliferation, and differentiation. Like all small GTPases, they work as molecular allosteric units that are active in signaling only when associated with the proper membrane compartment. Prenylation, occurring in the cytosol, is an enzymatic posttranslational event that anchors small GTPases at the membrane, and prenyl-binding proteins are needed to mask the cytoplasm-exposed lipid during transit to the target membrane. However, several of these proteins still await discovery. In this study, we report that cyclase-associated protein 1 (CAP1) binds Rap1. We found that this binding is GTP-independent, does not involve Rap1's effector domain, and is fully contained in its C-terminal hypervariable region (HVR). Furthermore, Rap1 prenylation was required for high-affinity interactions with CAP1 in a geranylgeranyl-specific manner. The prenyl binding specifically involved CAP1's C-terminal hydrophobic β-sheet domain. We present a combination of experimental and computational approaches, yielding a model whereby the high-affinity binding between Rap1 and CAP1 involves electrostatic and nonpolar side-chain interactions between Rap1's HVR residues, lipid, and CAP1 β-sheet domain. The binding was stabilized by the lipid insertion into the β-solenoid whose interior was occupied by nonpolar side chains. This model was reminiscent of the recently solved structure of the PDEδ-K-Ras complex; accordingly, disruptors of this complex, e.g. deltarasin, blocked the Rap1-CAP1 interaction. These findings indicate that CAP1 is a geranylgeranyl-binding partner of Rap1.
Keywords: CAP1; Ras proteins; Ras-related protein 1 (Rap1); cell signaling; chaperone; cyclase-associated protein 1; lipid-binding protein; molecular modeling; prenyl-binding protein; protein isoprenylation; small GTPase.
© 2018 Zhang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




Similar articles
-
CAP1 binds and activates adenylyl cyclase in mammalian cells.Proc Natl Acad Sci U S A. 2021 Jun 15;118(24):e2024576118. doi: 10.1073/pnas.2024576118. Proc Natl Acad Sci U S A. 2021. PMID: 34099549 Free PMC article.
-
The Cytoskeletal Protein Cyclase-Associated Protein 1 (CAP1) in Breast Cancer: Context-Dependent Roles in Both the Invasiveness and Proliferation of Cancer Cells and Underlying Cell Signals.Int J Mol Sci. 2019 May 30;20(11):2653. doi: 10.3390/ijms20112653. Int J Mol Sci. 2019. PMID: 31151140 Free PMC article. Review.
-
Characterization of interactions of adapter protein RAPL/Nore1B with RAP GTPases and their role in T cell migration.J Biol Chem. 2007 Oct 19;282(42):30629-42. doi: 10.1074/jbc.M704361200. Epub 2007 Aug 23. J Biol Chem. 2007. PMID: 17716979
-
Recognition and stabilization of geranylgeranylated human Rab5 by the GDP Dissociation Inhibitor (GDI).Small GTPases. 2019 May;10(3):227-242. doi: 10.1080/21541248.2017.1371268. Epub 2017 Oct 25. Small GTPases. 2019. PMID: 29065764 Free PMC article.
-
The Mystery of Rap1 Suppression of Oncogenic Ras.Trends Cancer. 2020 May;6(5):369-379. doi: 10.1016/j.trecan.2020.02.002. Epub 2020 Mar 2. Trends Cancer. 2020. PMID: 32249186 Free PMC article. Review.
Cited by
-
K-Ras G-domain binding with signaling lipid phosphatidylinositol (4,5)-phosphate (PIP2): membrane association, protein orientation, and function.J Biol Chem. 2019 Apr 26;294(17):7068-7084. doi: 10.1074/jbc.RA118.004021. Epub 2019 Feb 21. J Biol Chem. 2019. PMID: 30792310 Free PMC article.
-
Dynamic Phosphorylation and Dephosphorylation of Cyclase-Associated Protein 1 by Antagonistic Signaling through Cyclin-Dependent Kinase 5 and cAMP Are Critical for the Protein Functions in Actin Filament Disassembly and Cell Adhesion.Mol Cell Biol. 2020 Jan 30;40(4):e00282-19. doi: 10.1128/MCB.00282-19. Print 2020 Jan 30. Mol Cell Biol. 2020. PMID: 31791978 Free PMC article.
-
Phosphorylation promotes binding affinity of Rap-Raf complex by allosteric modulation of switch loop dynamics.Sci Rep. 2018 Aug 28;8(1):12976. doi: 10.1038/s41598-018-31234-7. Sci Rep. 2018. PMID: 30154518 Free PMC article.
-
Luminescence-activated nucleotide cyclase regulates spatial and temporal cAMP synthesis.J Biol Chem. 2019 Jan 25;294(4):1095-1103. doi: 10.1074/jbc.AC118.004905. Epub 2018 Dec 17. J Biol Chem. 2019. PMID: 30559293 Free PMC article.
-
Arl2-Mediated Allosteric Release of Farnesylated KRas4B from Shuttling Factor PDEδ.J Phys Chem B. 2018 Aug 2;122(30):7503-7513. doi: 10.1021/acs.jpcb.8b04347. Epub 2018 Jul 18. J Phys Chem B. 2018. PMID: 29961325 Free PMC article.
References
-
- Zhu L., Yang J., Bromberger T., Holly A., Lu F., Liu H., Sun K., Klapproth S., Hirbawi J., Byzova T. V., Plow E. F., Moser M., and Qin J. (2017) Structure of Rap1b bound to talin reveals a pathway for triggering integrin activation. Nat. Commun. 8, 1744 10.1038/s41467-017-01822-8 - DOI - PMC - PubMed
-
- Freeman S. A., Lei V., Dang-Lawson M., Mizuno K., Roskelley C. D., and Gold M. R. (2011) Cofilin-mediated F-actin severing is regulated by the Rap GTPase and controls the cytoskeletal dynamics that drive lymphocyte spreading and BCR microcluster formation. J. Immunol. 187, 5887–5900 10.4049/jimmunol.1102233 - DOI - PubMed
-
- Lin K. B., Freeman S. A., Zabetian S., Brugger H., Weber M., Lei V., Dang-Lawson M., Tse K. W., Santamaria R., Batista F. D., and Gold M. R. (2008) The rap GTPases regulate B cell morphology, immune-synapse formation, and signaling by particulate B cell receptor ligands. Immunity 28, 75–87 10.1016/j.immuni.2007.11.019 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

