The full-length cytochrome P450 enzyme CYP102A1 dimerizes at its reductase domains and has flexible heme domains for efficient catalysis
- PMID: 29618513
- PMCID: PMC5961045
- DOI: 10.1074/jbc.RA117.000600
The full-length cytochrome P450 enzyme CYP102A1 dimerizes at its reductase domains and has flexible heme domains for efficient catalysis
Abstract
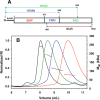
The cytochrome P450 enzyme CYP102A1 from Bacillus megaterium is a highly efficient hydroxylase of fatty acids, and there is a significant interest in using CYP102A1 for biotechnological applications. Here, we used size-exclusion chromatography-multiangle light scattering (SEC-MALS) analysis and negative-stain EM to investigate the molecular architecture of CYP102A1. The SEC-MALS analysis yielded a homogeneous peak with an average molecular mass of 235 ± 5 kDa, consistent with homodimeric CYP102A1. The negative-stain EM of dimeric CYP102A1 revealed four distinct lobes, representing the two heme and two reductase domains. Two of the lobes were in close contact, whereas the other two were often observed apart and at the ends of a U-shaped configuration. The overall dimension of the dimer was ∼130 Å. To determine the identity of the lobes, we FLAG-tagged the N or C terminus of CYP102A1 to visualize additional densities in EM and found that anti-FLAG Fab could bind only the N-tagged P450. Single-particle analysis of this anti-Flag Fab-CYP102A1 complex revealed additional density in the N-terminally tagged heme domains, indicating that the heme domains appear flexible, whereas the reductase domains remain tightly associated. The effects of truncation on CYP102A1 dimerization, identification of cross-linked sites by peptide mapping, and molecular modeling results all were consistent with the dimerization of the reductase domain. We conclude that functional CYP102A1 is a compact globular protein dimerized at its reductase domains, with its heme domains exhibiting multiple conformations that likely contribute to the highly efficient catalysis of CYP102A1.
Keywords: cytochrome P450; cytochrome P450 BM3; dimerization; electron microscopy (EM); electron transfer; fatty acid hydroxylase; heme domain; protein structure; reductase domain.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures






References
-
- Narhi L. O., and Fulco A. J. (1986) Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J. Biol. Chem. 261, 7160–7169 - PubMed
-
- Venkataraman H., Te Poele E. M., Rosloniec K. Z., Vermeulen N., Commandeur J. N., van der Geize R., and Dijkhuizen L. (2015) Biosynthesis of a steroid metabolite by an engineered Rhodococcus erythropolis strain expressing a mutant cytochrome P450 BM3 enzyme. Appl. Microbiol. Biotechnol. 99, 4713–4721 - PubMed
-
- Kim K. H., Kang J. Y., Kim D. H., Park S. H., Park S. H., Kim D., Park K. D., Lee Y. J., Jung H. C., Pan J. G., Ahn T., and Yun C. H. (2011) Generation of human chiral metabolites of simvastatin and lovastatin by bacterial CYP102A1 mutants. Drug Metab. Dispos. 39, 140–150 10.1124/dmd.110.036392 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

