The YTH Domain Protein ECT2 Is an m6A Reader Required for Normal Trichome Branching in Arabidopsis
- PMID: 29618631
- PMCID: PMC6002185
- DOI: 10.1105/tpc.17.00854
The YTH Domain Protein ECT2 Is an m6A Reader Required for Normal Trichome Branching in Arabidopsis
Abstract
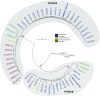
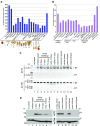
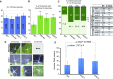
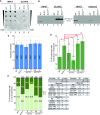
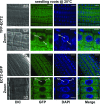
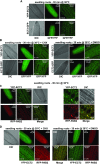
Methylations at position N6 of internal adenosines (m6As) are the most abundant and widespread mRNA modifications. These modifications play crucial roles in reproduction, growth, and development by controlling gene expression patterns at the posttranscriptional level. Their function is decoded by readers that share the YTH domain, which forms a hydrophobic pocket that directly accommodates the m6A residues. While the physiological and molecular functions of YTH readers have been extensively studied in animals, little is known about plant readers, even though m6As are crucial for plant survival and development. Viridiplantae contains high numbers of YTH domain proteins. Here, we performed comprehensive evolutionary analysis of YTH domain proteins and demonstrated that they are highly likely to be actual readers with redundant as well as specific functions. We also show that the ECT2 protein from Arabidopsis thaliana binds to m6A-containing RNAs in vivo and that this property relies on the m6A binding pocket carried by its YTH domain. ECT2 is cytoplasmic and relocates to stress granules upon heat exposure, suggesting that it controls mRNA fate in the cytosol. Finally, we demonstrate that ECT2 acts to decode the m6A signal in the trichome and is required for their normal branching through controlling their ploidy levels.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

