Linking Microbial Community Structure and Function During the Acidified Anaerobic Digestion of Grass
- PMID: 29619022
- PMCID: PMC5871674
- DOI: 10.3389/fmicb.2018.00540
Linking Microbial Community Structure and Function During the Acidified Anaerobic Digestion of Grass
Abstract
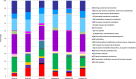
Harvesting valuable bioproducts from various renewable feedstocks is necessary for the critical development of a sustainable bioeconomy. Anaerobic digestion is a well-established technology for the conversion of wastewater and solid feedstocks to energy with the additional potential for production of process intermediates of high market values (e.g., carboxylates). In recent years, first-generation biofuels typically derived from food crops have been widely utilized as a renewable source of energy. The environmental and socioeconomic limitations of such strategy, however, have led to the development of second-generation biofuels utilizing, amongst other feedstocks, lignocellulosic biomass. In this context, the anaerobic digestion of perennial grass holds great promise for the conversion of sustainable renewable feedstock to energy and other process intermediates. The advancement of this technology however, and its implementation for industrial applications, relies on a greater understanding of the microbiome underpinning the process. To this end, microbial communities recovered from replicated anaerobic bioreactors digesting grass were analyzed. The bioreactors leachates were not buffered and acidic pH (between 5.5 and 6.3) prevailed at the time of sampling as a result of microbial activities. Community composition and transcriptionally active taxa were examined using 16S rRNA sequencing and microbial functions were investigated using metaproteomics. Bioreactor fraction, i.e., grass or leachate, was found to be the main discriminator of community analysis across the three molecular level of investigation (DNA, RNA, and proteins). Six taxa, namely Bacteroidia, Betaproteobacteria, Clostridia, Gammaproteobacteria, Methanomicrobia, and Negativicutes accounted for the large majority of the three datasets. The initial stages of grass hydrolysis were carried out by Bacteroidia, Gammaproteobacteria, and Negativicutes in the grass biofilms, in addition to Clostridia in the bioreactor leachates. Numerous glycolytic enzymes and carbohydrate transporters were detected throughout the bioreactors in addition to proteins involved in butanol and lactate production. Finally, evidence of the prevalence of stressful conditions within the bioreactors and particularly impacting Clostridia was observed in the metaproteomes. Taken together, this study highlights the functional importance of Clostridia during the anaerobic digestion of grass and thus research avenues allowing members of this taxon to thrive should be explored.
Keywords: 16S rRNA profiling; anaerobic digestion; biomolecule co-extraction; cellulosic substrate; metaproteomics.
Figures


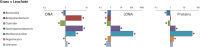



Similar articles
-
Effects of various feedstocks on isotope fractionation of biogas and microbial community structure during anaerobic digestion.Waste Manag. 2019 Feb 1;84:211-219. doi: 10.1016/j.wasman.2018.11.043. Epub 2018 Dec 4. Waste Manag. 2019. PMID: 30691895
-
Structural and functional analysis of the active cow rumen's microbial community provides a catalogue of genes and microbes participating in the deconstruction of cardoon biomass.Biotechnol Biofuels Bioprod. 2024 Apr 8;17(1):53. doi: 10.1186/s13068-024-02495-4. Biotechnol Biofuels Bioprod. 2024. PMID: 38589938 Free PMC article.
-
Recent advancements in strategies to improve anaerobic digestion of perennial energy grasses for enhanced methane production.Sci Total Environ. 2023 Feb 25;861:160552. doi: 10.1016/j.scitotenv.2022.160552. Epub 2022 Nov 28. Sci Total Environ. 2023. PMID: 36511320 Review.
-
Quantitative Metaproteomics Highlight the Metabolic Contributions of Uncultured Phylotypes in a Thermophilic Anaerobic Digester.Appl Environ Microbiol. 2016 Dec 30;83(2):e01955-16. doi: 10.1128/AEM.01955-16. Print 2017 Jan 15. Appl Environ Microbiol. 2016. PMID: 27815274 Free PMC article.
-
Metagenome, metatranscriptome, and metaproteome approaches unraveled compositions and functional relationships of microbial communities residing in biogas plants.Appl Microbiol Biotechnol. 2018 Jun;102(12):5045-5063. doi: 10.1007/s00253-018-8976-7. Epub 2018 Apr 30. Appl Microbiol Biotechnol. 2018. PMID: 29713790 Free PMC article. Review.
Cited by
-
Microbiome signature and diversity regulates the level of energy production under anaerobic condition.Sci Rep. 2021 Oct 5;11(1):19777. doi: 10.1038/s41598-021-99104-3. Sci Rep. 2021. PMID: 34611238 Free PMC article.
-
Enhanced anaerobic digestion of dairy wastewater in a granular activated carbon amended sequential batch reactor.Glob Change Biol Bioenergy. 2022 Jul;14(7):840-857. doi: 10.1111/gcbb.12947. Epub 2022 May 2. Glob Change Biol Bioenergy. 2022. PMID: 35915605 Free PMC article.
-
Emerging Strategies for Enhancing Propionate Conversion in Anaerobic Digestion: A Review.Molecules. 2023 May 4;28(9):3883. doi: 10.3390/molecules28093883. Molecules. 2023. PMID: 37175291 Free PMC article. Review.
-
Convergent Microbial Community Formation in Replicate Anaerobic Reactors Inoculated from Different Sources and Treating Ersatz Crew Waste.Life (Basel). 2021 Dec 10;11(12):1374. doi: 10.3390/life11121374. Life (Basel). 2021. PMID: 34947905 Free PMC article.
-
A bibliometric analysis of the global impact of metaproteomics research.Front Microbiol. 2023 Jul 5;14:1217727. doi: 10.3389/fmicb.2023.1217727. eCollection 2023. Front Microbiol. 2023. PMID: 37476667 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

