Immunogenicity of a Fap2 peptide mimotope of Fusobacterium nucleatum and its potential use in the diagnosis of colorectal cancer
- PMID: 29619076
- PMCID: PMC5879760
- DOI: 10.1186/s13027-018-0184-7
Immunogenicity of a Fap2 peptide mimotope of Fusobacterium nucleatum and its potential use in the diagnosis of colorectal cancer
Abstract
Background: The role of Fusobacterium nucleatum Fap2 protein in the development of colorectal cancer has recently been explained. Fap2, when bound to the human inhibitory receptor, TIGIT, inhibits the cytotoxic activity of natural killer (NK) cells against cancer cells, thus, allowing proliferation of the latter eventually leading to tumor growth. The aim of the study was to identify the immunogenicity of a peptide mimotope of the Fap2 protein and to determine the reactivity of colorectal cancer patients' sera against the mimotope.
Methods: Immunogenic epitope of the Fap2 protein of F. nucleatum was selected using the B-cell epitope prediction of the Immune Epitope Database and Analysis Resource (IEDB). The immunogenicity of the synthetic peptide mimotope of the Fap2 protein was determined in animal models and reactivity of colorectal cancer patients' sera against the mimotope was done by indirect ELISA.
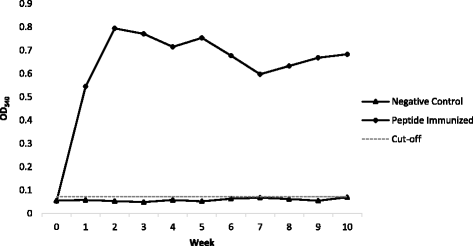
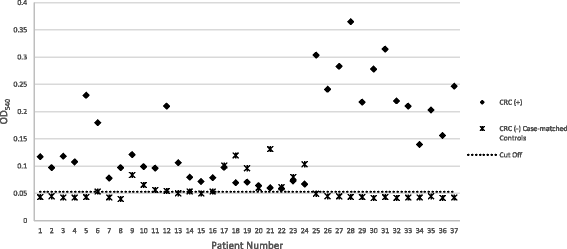
Results: Results show that the selected peptide mimotope, with sequence TELAYKHYFGT, of the outer membrane protein Fap2 of F. nucleatum is immunogenic. Increase in the absorbance readings of peptide-immunized rabbit sera was observed starting Week 1 which was sustained up to Week 10 in the indirect ELISA performed. Colorectal cancer cases (n = 37) were all reactive in an ELISA-based analysis using the mimotope as the capture antigen.
Conclusions: In this study, we identified an immunogenic epitope of the Fap2 protein of the Fusobacterium nucleatum. We demonstrated the reactivity of serum of histopathologically confirmed CRC patients in a peptide-capture indirect ELISA which may serve as proof of concept for the development of CRC diagnostics.
Keywords: Colorectal cancer; ELISA; Fap2 protein; Fusobacterium nucleatum; Immunodiagnostics.
Conflict of interest statement
Mr. Leonardo A. Guevarra Jr., the principal investigator of this project and the corresponding author for this study, is a faculty member at the Department of Biochemistry, Faculty of Pharmacy and Research Faculty at the Research Center for Natural and Applied Sciences, University of Santo Tomas. He first got his interest in infectious disease, cancer, and diagnostics development when he was doing his research for the Master of Science in Biochemistry at the Department of Biochemistry and Molecular Biology, College of Medicine, University of the Philippines - Manila. His current organizational involvement includes the Philippine Society of Biochemistry and Molecular Biology (lifetime member), UK Biochemical Society, and the Japanese Peptide Society where he recently presented (poster presentation) the results in the paper being submitted for publication. Mr. Guevarra is establishing his research career in immune epitope prediction and its application in infectious disease and cancer diagnostics.This study was approved by the Institutional Review Board of the University of Santo Tomas Hospital chaired by Dr. Wilson L. Tan De Guzman. You can contact the IRB by email at ust_irb@yahoo.com.ph. Written informed consent for the use of leftover serum collected from routine serological testing was requested from patients participating in the study. Clearance and approval from the Institutional Animal Care and Utilization Committee of the University of Santo Tomas was acquired. The UST IACUC is chaired by Prof. Jovencio G. Apostol, PhD and can be contacted through ustrcns@yahoo.com.Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Bakken V, Høgh BT, Jensen HB. Growth conditions and outer membrane proteins of Fusobacterium nucleatum. Scand J Dent Res. 1990;98:215–224. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

