The Role of Intestinal C-type Regenerating Islet Derived-3 Lectins for Nonalcoholic Steatohepatitis
- PMID: 29619418
- PMCID: PMC5880191
- DOI: 10.1002/hep4.1165
The Role of Intestinal C-type Regenerating Islet Derived-3 Lectins for Nonalcoholic Steatohepatitis
Abstract
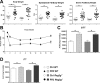
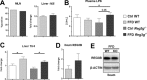
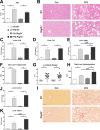
C-type regenerating islet derived-3 (Reg3) lectins defend against pathogens and keep commensal bacteria at a distance. Deficiency of Reg3g and Reg3b facilitates alcohol-induced bacterial translocation and alcoholic liver disease. Intestinal Reg3g is down-regulated in animal models of diet-induced obesity, but the functional consequences for nonalcoholic steatohepatitis (NASH) are unknown. The aim of this study was to investigate the role of Reg3 lectins in NASH. NASH was induced by a Western-style fast-food diet in mice deficient for Reg3g or Reg3b and in transgenic mice overexpressing Reg3g in intestinal epithelial cells (Reg3gTg). Glucose tolerance was assessed after 18 weeks and insulin resistance after 19 weeks of feeding. After 20 weeks, mice were assessed for features of the metabolic syndrome. Obesity was not different in genetically modified mice compared with their respective wild-type littermates. Glucose intolerance, liver injury, hepatic inflammation, steatosis, fibrosis, and bacterial translocation to mesenteric lymph nodes and to the liver were not different in Reg3g-deficient mice compared with wild-type littermates. Plasma endotoxin levels were higher in Reg3g-deficient mice. Reg3b deficiency protected against glucose intolerance, but liver disease, bacterial translocation, and plasma endotoxin levels were similar to wild-type littermates. Absence of either REG3G or REG3B protein in the ileum was not compensated for by up-regulation of the respective other REG3 protein. Transgenic Reg3g mice also developed liver injury, steatosis, and fibrosis similar to their wild-type littermates. Conclusion: In contrast to alcoholic liver disease, loss of intestinal Reg3 lectins is not sufficient to aggravate diet-induced obesity and NASH. This supports a multi-hit pathogenesis in NASH. Only glucose metabolism is affected by Reg3b deficiency. (Hepatology Communications 2018;2:393-406).
Figures





References
-
- Diehl AM, Day C. Cause, pathogenesis, and treatment of nonalcoholic steatohepatitis. N Engl J Med 2017;377:2063‐2072. - PubMed
-
- Llopis M, Cassard AM, Wrzosek L, Boschat L, Bruneau A, Ferrere G, et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut 2016;65:830‐839. - PubMed
-
- Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, et al. Intestinal microbiota determines development of non‐alcoholic fatty liver disease in mice. Gut 2013;62:1787‐1794. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

