Exploring the molecular mechanisms of osteosarcoma by the integrated analysis of mRNAs and miRNA microarrays
- PMID: 29620143
- PMCID: PMC5979835
- DOI: 10.3892/ijmm.2018.3594
Exploring the molecular mechanisms of osteosarcoma by the integrated analysis of mRNAs and miRNA microarrays
Abstract
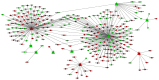
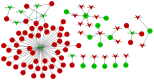
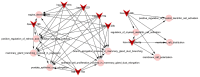
Osteosarcoma (OS) is the most frequently occurring primary bone malignancy with a rapid progression and poor survival. In the present study, in order to examine the molecular mechanisms of OS, we analyzed the microarray of GSE28425. GSE28425 was downloaded from Gene Expression Omnibus, which also included the miRNA expression profile, GSE28423, and the mRNA expression profile, GSE28424. Each of the expression profiles included 19 OS cell lines and 4 normal bones. The differentially expressed genes (DEGs) and differentially expressed miRNAs (DE-miRNAs) were screened using the limma package in Bioconductor. The DEGs associated with tumors were screened and annotated. Subsequently, the potential functions of the DEGs were analyzed by Gene Ontology (GO) and pathway enrichment analyses. Furthermore, the protein-protein interaction (PPI) network was constructed using the STRING database and Cytoscape software. Furthermore, modules of the PPI network were screened using the ClusterOne plugin in Cytoscape. Additionally, the transcription factor (TF)-DEG regulatory network, DE-miRNA-DEG regulatory network and miRNA-function collaborative network were separately constructed to obtain key DEGs and DE-miRNAs. In total, 1,609 DEGs and 149 DE-miRNAs were screened. Upregulated FOS-like antigen 1 (FOSL1) also had the function of an oncogene. MAD2 mitotic arrest deficient-like 1 (MAD2L1; degree, 65) and aurora kinase A (AURKA; degree, 64) had higher degrees in the PPI network of the DEGs. In the TF-DEG regulatory network, the TF, signal transducer and activator of transcription 3 (STAT3) targeted the most DEGs. Moreover, in the DE-miRNA-DEG regulatory network, downregulated miR‑1 targeted many DEGs and estrogen receptor 1 (ESR1) was targeted by several highly expressed miRNAs. Moreover, in the miRNA-function collaborative networks of upregulated miRNAs, miR‑128 targeted myeloid dendritic associated functions. On the whole, our data indicate that MAD2L1, AURKA, STAT3, ESR1, FOSL1, miR‑1 and miR‑128 may play a role in the development and/or progressio of OS.
Figures


References
-
- Wittig JC, Bickels J, Priebat D, Jelinek J, Kellar-Graney K, Shmookler B, Malawer MM. Osteosarcoma: A multidisciplinary approach to diagnosis and treatment. Am Fam Physician. 2002;65:1123–1132. - PubMed
-
- Ottaviani G, Jaffe N. Pediatric and Adolescent Osteosarcoma. Springer; 2010. The epidemiology of osteosarcoma; pp. 3–13.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

