Long noncoding RNA LINC01296 promotes tumor growth and progression by sponging miR-5095 in human cholangiocarcinoma
- PMID: 29620172
- PMCID: PMC5919714
- DOI: 10.3892/ijo.2018.4362
Long noncoding RNA LINC01296 promotes tumor growth and progression by sponging miR-5095 in human cholangiocarcinoma
Abstract
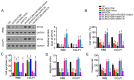
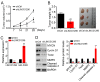
The aim of the present study was to elucidate whether, and how, long intergenic non-protein coding RNA 1296 (LINC01296) is involved in the modulation of human cholangiocarcinoma (CCA) development and progression. Microarray data analysis and reverse transcription-quantitative polymerase chain reaction analysis demonstrated that LINC01296 was significantly upregulated in human CCA compared with nontumor tissues. Furthermore, the expression of LINC01296 in human CCA was positively associated with tumor severity and clinical stage. Knockdown of LINC01296 dramatically suppressed the viability, migration and invasion of RBE and CCLP1 cells, and promoted cell apoptosis in vitro. Furthermore, LINC01296 knockdown inhibited tumor growth in a xenograft model. Mechanistically, LINC01296 was demonstrated to sponge microRNA-5095 (miR-5095), which targets MYCN proto-oncogene bHLH transcription factor (MYCN) mRNA in human CCA. By inhibition of miR-5095, LINC01296 overexpression upregulated the expression of MYCN and promoted cell viability, migration and invasion in CCA cells. The results reveal that the axis of LINC01296/miR-5095/MYCN may be a mechanism to regulate CCA development and progression.
Figures





References
-
- Welzel TM, Graubard BI, El-Serag HB, Shaib YH, Hsing AW, Davila JA, McGlynn KA. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma in the United States: A population-based case-control study. Clin Gastroenterol Hepatol. 2007;5:1221–1228. doi: 10.1016/j.cgh.2007.05.020. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

