Translational control of the undifferentiated phenotype in ER‑positive breast tumor cells: Cytoplasmic localization of ERα and impact of IRES inhibition
- PMID: 29620220
- PMCID: PMC5983923
- DOI: 10.3892/or.2018.6332
Translational control of the undifferentiated phenotype in ER‑positive breast tumor cells: Cytoplasmic localization of ERα and impact of IRES inhibition
Abstract
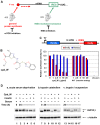
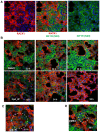
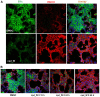
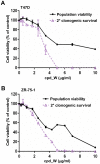
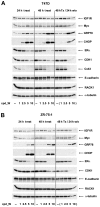
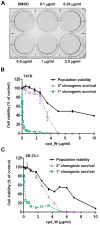
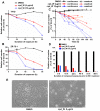
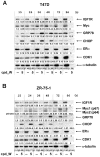
Using a series of potential biomarkers relevant to mechanisms of protein synthesis, we observed that estrogen receptor (ER)-positive breast tumor cells exist in two distinct yet interconvertible phenotypic states (of roughly equal proportion) which differ in the degree of differentiation and use of IRES-mediated translation. Nascently translated IGF1R in the cytoplasm positively correlated with IRES activity and the undifferentiated phenotype, while epitope accessibility of RACK1, an integral component of the 40S ribosomal subunit, aligned with the more differentiated IRES-off state. When deprived of soluble growth factors, the entire tumor cell population shifted to the undifferentiated phenotype in which IRES-mediated translation was active, facilitating survival under these adverse microenvironmental conditions. However, if IRES-mediated translation was inhibited, the cells instead were forced to transition uniformly to the more differentiated state. Notably, cytoplasmic localization of estrogen receptor α (ERα/ESR1) precisely mirrored the pattern observed with nascent IGF1R, correlating with the undifferentiated IRES-active phenotype. Inhibition of IRES-mediated translation resulted in both a shift in ERα to the nucleus (consistent with differentiation) and a marked decrease in ERα abundance (consistent with the inhibition of ERα synthesis via its IRES). Although breast tumor cells tolerated forced differentiation without extensive loss of their viability, their reproductive capacity was severely compromised. In addition, CDK1 was decreased, connexin 43 eliminated and Myc translation altered as a consequence of IRES inhibition. Isolated or low-density ER-positive breast tumor cells were particularly vulnerable to IRES inhibition, losing the ability to generate viable cohesive colonies, or undergoing massive cell death. Collectively, these results provide further evidence for the integral relationship between IRES-mediated translation and the undifferentiated phenotype and demonstrate how therapeutic manipulation of this specialized mode of protein synthesis may be used to limit the phenotypic plasticity and incapacitate or eliminate these otherwise highly resilient breast tumor cells.
Figures


Similar articles
-
IRES inhibition induces terminal differentiation and synchronized death in triple-negative breast cancer and glioblastoma cells.Tumour Biol. 2016 Oct;37(10):13247-13264. doi: 10.1007/s13277-016-5161-4. Epub 2016 Jul 26. Tumour Biol. 2016. PMID: 27460074 Free PMC article.
-
Northwestern profiling of potential translation-regulatory proteins in human breast epithelial cells and malignant breast tissues: evidence for pathological activation of the IGF1R IRES.Exp Mol Pathol. 2010 Jun;88(3):341-52. doi: 10.1016/j.yexmp.2010.03.006. Epub 2010 Mar 15. Exp Mol Pathol. 2010. PMID: 20233590 Free PMC article.
-
Upregulation of IRS1 Enhances IGF1 Response in Y537S and D538G ESR1 Mutant Breast Cancer Cells.Endocrinology. 2018 Jan 1;159(1):285-296. doi: 10.1210/en.2017-00693. Endocrinology. 2018. PMID: 29029116 Free PMC article.
-
Small molecule inhibitors of IRES-mediated translation.Cancer Biol Ther. 2015;16(10):1471-85. doi: 10.1080/15384047.2015.1071729. Epub 2015 Jul 15. Cancer Biol Ther. 2015. PMID: 26177060 Free PMC article.
-
Hepatitis C Virus Translation Regulation.Int J Mol Sci. 2020 Mar 27;21(7):2328. doi: 10.3390/ijms21072328. Int J Mol Sci. 2020. PMID: 32230899 Free PMC article. Review.
Cited by
-
IRES Trans-Acting Factors, Key Actors of the Stress Response.Int J Mol Sci. 2019 Feb 20;20(4):924. doi: 10.3390/ijms20040924. Int J Mol Sci. 2019. PMID: 30791615 Free PMC article. Review.
-
Isoform-Directed Control of c-Myc Functions: Understanding the Balance from Proliferation to Growth Arrest.Int J Mol Sci. 2023 Dec 15;24(24):17524. doi: 10.3390/ijms242417524. Int J Mol Sci. 2023. PMID: 38139353 Free PMC article. Review.
References
-
- Meng Z, Jackson NL, Choi H, King PH, Emanuel PD, Blume SW. Alterations in RNA-binding activities of IRES-regulatory proteins as a mechanism for physiological variability and pathological dysregulation of IGF-IR translational control in human breast tumor cells. J Cell Physiol. 2008;217:172–183. doi: 10.1002/jcp.21486. - DOI - PubMed
-
- Nagamachi A, Htun PW, Ma F, Miyazaki K, Yamasaki N, Kanno M, Inaba T, Honda Z, Okuda T, Oda H, et al. A 5′untranslated region containing the IRES element in the Runx1 gene is required for angiogenesis, hematopoiesis and leukemogenesis in a knock-in mouse model. Dev Biol. 2010;345:226–236. doi: 10.1016/j.ydbio.2010.07.015. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

