Applications of CRISPR/Cas System to Bacterial Metabolic Engineering
- PMID: 29621180
- PMCID: PMC5979482
- DOI: 10.3390/ijms19041089
Applications of CRISPR/Cas System to Bacterial Metabolic Engineering
Abstract
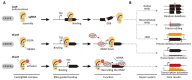
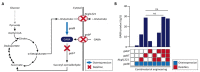
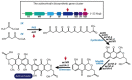
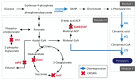
The clustered regularly interspaced short palindromic repeats/CRISPR-associated (CRISPR/Cas) adaptive immune system has been extensively used for gene editing, including gene deletion, insertion, and replacement in bacterial and eukaryotic cells owing to its simple, rapid, and efficient activities in unprecedented resolution. Furthermore, the CRISPR interference (CRISPRi) system including deactivated Cas9 (dCas9) with inactivated endonuclease activity has been further investigated for regulation of the target gene transiently or constitutively, avoiding cell death by disruption of genome. This review discusses the applications of CRISPR/Cas for genome editing in various bacterial systems and their applications. In particular, CRISPR technology has been used for the production of metabolites of high industrial significance, including biochemical, biofuel, and pharmaceutical products/precursors in bacteria. Here, we focus on methods to increase the productivity and yield/titer scan by controlling metabolic flux through individual or combinatorial use of CRISPR/Cas and CRISPRi systems with introduction of synthetic pathway in industrially common bacteria including Escherichia coli. Further, we discuss additional useful applications of the CRISPR/Cas system, including its use in functional genomics.
Keywords: CRISPR/Cas; CRISPRa; CRISPRi; gene regulation; genome editing; metabolic engineering.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

