A genomic overview of the population structure of Salmonella
- PMID: 29621240
- PMCID: PMC5886390
- DOI: 10.1371/journal.pgen.1007261
A genomic overview of the population structure of Salmonella
Abstract
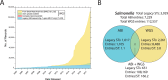
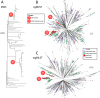
For many decades, Salmonella enterica has been subdivided by serological properties into serovars or further subdivided for epidemiological tracing by a variety of diagnostic tests with higher resolution. Recently, it has been proposed that so-called eBurst groups (eBGs) based on the alleles of seven housekeeping genes (legacy multilocus sequence typing [MLST]) corresponded to natural populations and could replace serotyping. However, this approach lacks the resolution needed for epidemiological tracing and the existence of natural populations had not been independently validated by independent criteria. Here, we describe EnteroBase, a web-based platform that assembles draft genomes from Illumina short reads in the public domain or that are uploaded by users. EnteroBase implements legacy MLST as well as ribosomal gene MLST (rMLST), core genome MLST (cgMLST), and whole genome MLST (wgMLST) and currently contains over 100,000 assembled genomes from Salmonella. It also provides graphical tools for visual interrogation of these genotypes and those based on core single nucleotide polymorphisms (SNPs). eBGs based on legacy MLST are largely consistent with eBGs based on rMLST, thus demonstrating that these correspond to natural populations. rMLST also facilitated the selection of representative genotypes for SNP analyses of the entire breadth of diversity within Salmonella. In contrast, cgMLST provides the resolution needed for epidemiological investigations. These observations show that genomic genotyping, with the assistance of EnteroBase, can be applied at all levels of diversity within the Salmonella genus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Gwyn LB (1898) On infection with a Para-Colin bacillus in a case with all the clinical features of typhoid fever. Johns Hopkins Hospital Bulletin 84: 54–56.
-
- Grimont P. A. and Weill F.-X. (2007) Antigenic formulae of the Salmonella serovars Paris, France: WHO Collaborating Centre for Reference and Research on Salmonella. 166 p.
-
- Achtman M, Wain J, Weill F-X, Nair S, Zhou Z, Sangal V, et al. (2012) Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog 8: e1002776 doi: 10.1371/journal.ppat.1002776 - DOI - PMC - PubMed
-
- Kidgell C, Reichard U, Wain J, Linz B, Torpdahl M, Dougan G, et al. (2002) Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect Genet Evol 2: 39–45. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

