Repression of Nitrogen Starvation Responses by Members of the Arabidopsis GARP-Type Transcription Factor NIGT1/HRS1 Subfamily
- PMID: 29622567
- PMCID: PMC5969275
- DOI: 10.1105/tpc.17.00810
Repression of Nitrogen Starvation Responses by Members of the Arabidopsis GARP-Type Transcription Factor NIGT1/HRS1 Subfamily
Abstract
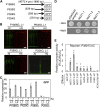
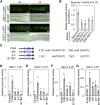
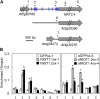
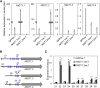
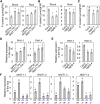
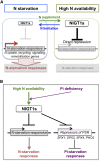
Nitrogen (N) is often a limiting nutrient whose availability determines plant growth and productivity. Because its availability is often low and/or not uniform over time and space in nature, plants respond to variations in N availability by altering uptake and recycling mechanisms, but the molecular mechanisms underlying how these responses are regulated are poorly understood. Here, we show that a group of GARP G2-like transcription factors, Arabidopsis thaliana NITRATE-INDUCIBLE, GARP-TYPE TRANSCRIPTIONAL REPRESSOR1/HYPERSENSITIVE TO LOW Pi-ELICITED PRIMARY ROOT SHORTENING1 proteins (NIGT1/HRS1s), are factors that bind to the promoter of the N starvation marker NRT2.4 and repress an array of N starvation-responsive genes under conditions of high N availability. Transient assays and expression analysis demonstrated that NIGT1/HRS1s are transcriptional repressors whose expression is regulated by N availability. We identified target genes of the NIGT1/HRS1s by genome-wide transcriptome analyses and found that they are significantly enriched in N starvation response-related genes, including N acquisition, recycling, remobilization, and signaling genes. Loss of NIGT1/HRS1s resulted in deregulation of N acquisition and accumulation. We propose that NIGT1/HRS1s are major regulators of N starvation responses that play an important role in optimizing N acquisition and utilization under fluctuating N conditions.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures







References
-
- Araya T., Miyamoto M., Wibowo J., Suzuki A., Kojima S., Tsuchiya Y.N., Sawa S., Fukuda H., von Wirén N., Takahashi H. (2014). CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. USA 111: 2029–2034. - PMC - PubMed
-
- Avila-Ospina L., Moison M., Yoshimoto K., Masclaux-Daubresse C. (2014). Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 65: 3799–3811. - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300.
-
- Camañes G., Bellmunt E., García-Andrade J., García-Agustín P., Cerezo M. (2012). Reciprocal regulation between AtNRT2.1 and AtAMT1.1 expression and the kinetics of NH4+ and NO3- influxes. J. Plant Physiol. 169: 268–274. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

