Coordination of Protein Kinase and Phosphoprotein Phosphatase Activities in Mitosis
- PMID: 29623276
- PMCID: PMC5874294
- DOI: 10.3389/fcell.2018.00030
Coordination of Protein Kinase and Phosphoprotein Phosphatase Activities in Mitosis
Abstract
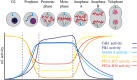
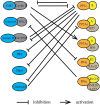
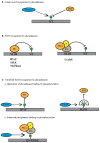
Dynamic changes in protein phosphorylation govern the transitions between different phases of the cell division cycle. A "tug of war" between highly conserved protein kinases and the family of phosphoprotein phosphatases (PPP) establishes the phosphorylation state of proteins, which controls their function. More than three-quarters of all proteins are phosphorylated at one or more sites in human cells, with the highest occupancy of phosphorylation sites seen in mitosis. Spatial and temporal regulation of opposing kinase and PPP activities is crucial for accurate execution of the mitotic program. The role of mitotic kinases has been the focus of many studies, while the contribution of PPPs was for a long time underappreciated and is just emerging. Misconceptions regarding the specificity and activity of protein phosphatases led to the belief that protein kinases are the primary determinants of mitotic regulation, leaving PPPs out of the limelight. Recent studies have shown that protein phosphatases are specific and selective enzymes, and that their activity is tightly regulated. In this review, we discuss the emerging roles of PPPs in mitosis and their regulation of and by mitotic kinases, as well as mechanisms that determine PPP substrate recognition and specificity.
Keywords: kinases and phosphatase; mass spectrometry; mitosis; phosphatases; protoemics.
Figures
References
-
- Álvarez-Fernández M., Sánchez-Martínez R., Sanz-Castillo B., Gan P. P., Sanz-Flores M., Malumbres M., et al. . (2013). Greatwall is essential to prevent mitotic collapse after nuclear envelope breakdown in mammals. Proc. Natl. Acad. Sci. U.S.A. 110, 17374–17379. 10.1073/pnas.1310745110 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

