Identification of PP1-Gadd34 substrates involved in the unfolded protein response using K-BIPS, a method for phosphatase substrate identification
- PMID: 29623310
- PMCID: PMC5997272
- DOI: 10.1039/c7mo00064b
Identification of PP1-Gadd34 substrates involved in the unfolded protein response using K-BIPS, a method for phosphatase substrate identification
Abstract
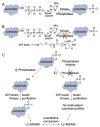
Phosphorylation is a key post-translational modification in cell signaling, which is regulated by the equilibrium activities of kinases and phosphatases. The biological significance of many phosphorylation events remains poorly characterized due to the scarcity of tools to discover phosphatases substrates. In prior work, we established kinase-catalyzed biotinylation where kinases accept the γ-modified ATP analog, ATP-biotin, to label phosphoproteins. Here, we developed a novel method to study substrates of phosphatases using kinase-catalyzed biotinylation termed K-BIPS (Kinase-catalyzed Biotinylation to Identify Phosphatase Substrates). In a proof-of-concept experiment, K-BIPS was initially used to explore the substrates of phosphatases inhibited by okadaic acid. Many known phosphatase substrates were observed, confirming K-BIPS as a valid phosphatase substrate identification tool. Then, as a further application, K-BIPS was used to discover the substrates of the PP1-Gadd34 phosphatase complex in the context of unfolded protein response (UPR). In addition to the known substrate eIF2α, K-BIPS revealed several novel substrates, suggesting a more prominent role for the PP1-Gadd34 complex in UPR than previously appreciated. Overall, the two studies establish K-BIPS as a powerful tool to discover the cellular substrates of phosphatases.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Kinase-Catalyzed Biotinylation to Identify Phosphatase Substrates (K-BIPS).Methods Mol Biol. 2024;2743:135-152. doi: 10.1007/978-1-0716-3569-8_9. Methods Mol Biol. 2024. PMID: 38147213 Free PMC article.
-
l-Lactate Dehydrogenase Identified as a Protein Tyrosine Phosphatase 1B Substrate by Using K-BIPS.Chembiochem. 2021 Jan 5;22(1):186-192. doi: 10.1002/cbic.202000499. Epub 2020 Dec 7. Chembiochem. 2021. PMID: 33002308 Free PMC article.
-
Biotinylated phosphoproteins from kinase-catalyzed biotinylation are stable to phosphatases: implications for phosphoproteomics.Chembiochem. 2013 Feb 11;14(3):381-7. doi: 10.1002/cbic.201200626. Epub 2013 Jan 17. Chembiochem. 2013. PMID: 23335220 Free PMC article.
-
Roles of protein kinases and phosphatases in signal transduction.Symp Soc Exp Biol. 1990;44:241-55. Symp Soc Exp Biol. 1990. PMID: 1966636 Review.
-
Function and regulation of serine/threonine phosphatases in the healthy and diseased heart.J Mol Cell Cardiol. 2013 Nov;64:90-8. doi: 10.1016/j.yjmcc.2013.09.006. Epub 2013 Sep 16. J Mol Cell Cardiol. 2013. PMID: 24051368 Review.
Cited by
-
Identification of PP1c-PPP1R12A Substrates Using Kinase-Catalyzed Biotinylation to Identify Phosphatase Substrates.ACS Omega. 2023 Sep 21;8(39):35628-35637. doi: 10.1021/acsomega.3c01944. eCollection 2023 Oct 3. ACS Omega. 2023. PMID: 37810667 Free PMC article.
-
The endoplasmic reticulum: Homeostasis and crosstalk in retinal health and disease.Prog Retin Eye Res. 2024 Jan;98:101231. doi: 10.1016/j.preteyeres.2023.101231. Epub 2023 Dec 12. Prog Retin Eye Res. 2024. PMID: 38092262 Free PMC article. Review.
-
The integrated stress response: From mechanism to disease.Science. 2020 Apr 24;368(6489):eaat5314. doi: 10.1126/science.aat5314. Science. 2020. PMID: 32327570 Free PMC article. Review.
-
Kinase-Catalyzed Biotinylation to Identify Phosphatase Substrates (K-BIPS).Methods Mol Biol. 2024;2743:135-152. doi: 10.1007/978-1-0716-3569-8_9. Methods Mol Biol. 2024. PMID: 38147213 Free PMC article.
-
Delineating the role of eIF2α in retinal degeneration.Cell Death Dis. 2019 May 28;10(6):409. doi: 10.1038/s41419-019-1641-y. Cell Death Dis. 2019. PMID: 31138784 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

