Oncogenic Signaling Pathways in The Cancer Genome Atlas
- PMID: 29625050
- PMCID: PMC6070353
- DOI: 10.1016/j.cell.2018.03.035
Oncogenic Signaling Pathways in The Cancer Genome Atlas
Abstract
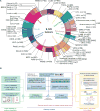
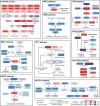
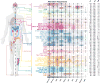
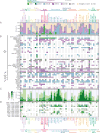
Genetic alterations in signaling pathways that control cell-cycle progression, apoptosis, and cell growth are common hallmarks of cancer, but the extent, mechanisms, and co-occurrence of alterations in these pathways differ between individual tumors and tumor types. Using mutations, copy-number changes, mRNA expression, gene fusions and DNA methylation in 9,125 tumors profiled by The Cancer Genome Atlas (TCGA), we analyzed the mechanisms and patterns of somatic alterations in ten canonical pathways: cell cycle, Hippo, Myc, Notch, Nrf2, PI-3-Kinase/Akt, RTK-RAS, TGFβ signaling, p53 and β-catenin/Wnt. We charted the detailed landscape of pathway alterations in 33 cancer types, stratified into 64 subtypes, and identified patterns of co-occurrence and mutual exclusivity. Eighty-nine percent of tumors had at least one driver alteration in these pathways, and 57% percent of tumors had at least one alteration potentially targetable by currently available drugs. Thirty percent of tumors had multiple targetable alterations, indicating opportunities for combination therapy.
Keywords: PanCanAtlas; TCGA; cancer genome atlas; cancer genomics; combination therapy; pan-cancer; precision oncology; signaling pathways; therapeutics; whole exome sequencing.
Copyright © 2018. Published by Elsevier Inc.
Figures



Comment in
-
The Pan-Cancer Atlas: a New Chapter in Cancer Molecular Targeting Therapy.Pathol Oncol Res. 2020 Jul;26(3):1997-1999. doi: 10.1007/s12253-019-00709-x. Epub 2019 Aug 29. Pathol Oncol Res. 2020. PMID: 31468361 No abstract available.
References
-
- Aster JC, Blacklow SC. Targeting the Notch pathway: twists and turns on the road to rational therapeutics. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2012;30:2418–2420. - PubMed
-
- Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Betrand D, Weerasinghe A, Colaprico A, Kim J, Wendl MC, Reardon B, et al. Comprehensive Discovery and Characterization of Driver Genes and Mutations in Human Cancers. Cell 2017
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- U24 CA143882/CA/NCI NIH HHS/United States
- U24 CA143866/CA/NCI NIH HHS/United States
- P41 GM103504/GM/NIGMS NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- U24 CA143840/CA/NCI NIH HHS/United States
- U24 CA143858/CA/NCI NIH HHS/United States
- U24 CA143848/CA/NCI NIH HHS/United States
- U24 CA210949/CA/NCI NIH HHS/United States
- R01 CA163722/CA/NCI NIH HHS/United States
- T32 HG000046/HG/NHGRI NIH HHS/United States
- U24 CA143867/CA/NCI NIH HHS/United States
- U24 CA210990/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U24 CA143835/CA/NCI NIH HHS/United States
- U24 CA210950/CA/NCI NIH HHS/United States
- U24 CA143845/CA/NCI NIH HHS/United States
- U24 CA143799/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U24 CA144025/CA/NCI NIH HHS/United States
- U24 CA143843/CA/NCI NIH HHS/United States
- T32 CA009676/CA/NCI NIH HHS/United States
- P30 ES013508/ES/NIEHS NIH HHS/United States
- U24 CA210957/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- U24 CA210969/CA/NCI NIH HHS/United States
- U24 CA143883/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

