A nutrient-dependent division antagonist is regulated post-translationally by the Clp proteases in Bacillus subtilis
- PMID: 29625553
- PMCID: PMC5889556
- DOI: 10.1186/s12866-018-1155-2
A nutrient-dependent division antagonist is regulated post-translationally by the Clp proteases in Bacillus subtilis
Abstract
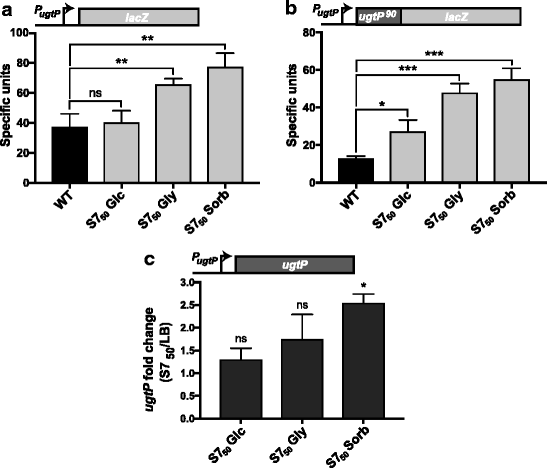
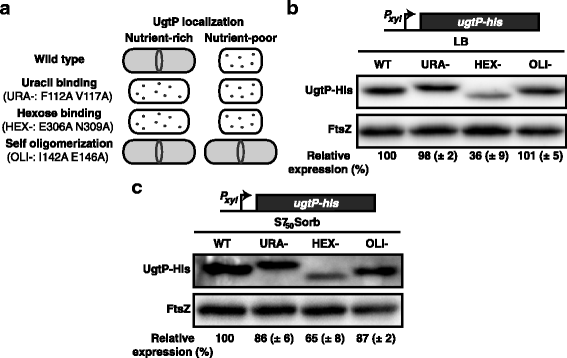
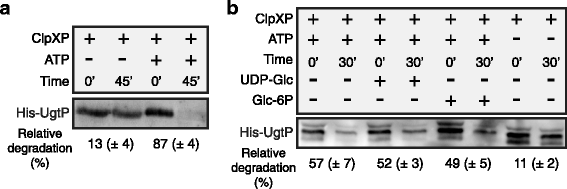
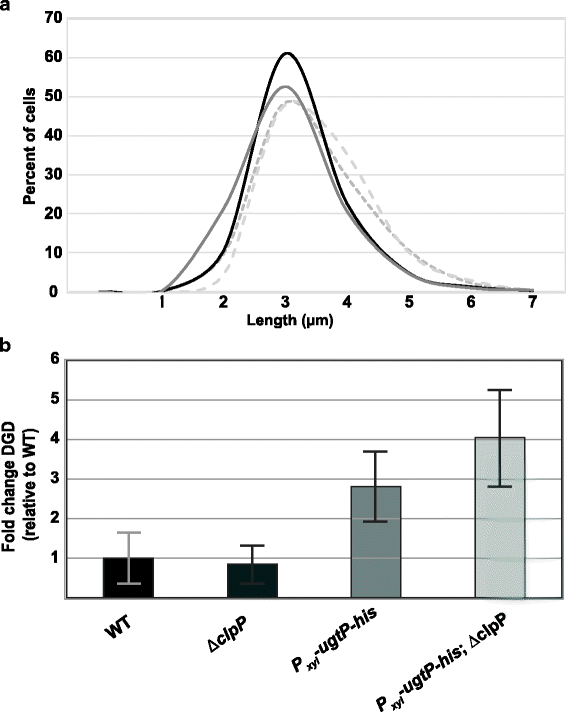
Background: Changes in nutrient availability have dramatic and well-defined impacts on both transcription and translation in bacterial cells. At the same time, the role of post-translational control in adaptation to nutrient-poor environments is poorly understood. Previous studies demonstrate the ability of the glucosyltransferase UgtP to influence cell size in response to nutrient availability. Under nutrient-rich medium, interactions with its substrate UDP-glucose promote interactions between UgtP and the tubulin-like cell division protein FtsZ in Bacillus subtilis, inhibiting maturation of the cytokinetic ring and increasing cell size. In nutrient-poor medium, reductions in UDP-glucose availability favor UgtP oligomerization, sequestering it from FtsZ and allowing division to occur at a smaller cell mass.
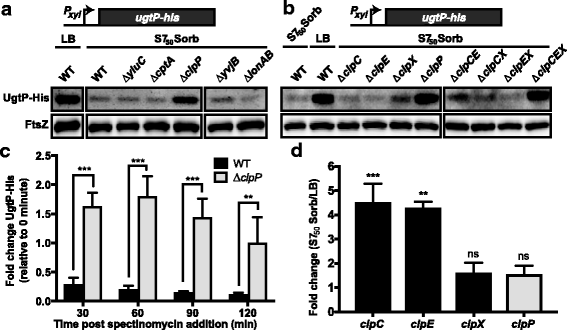
Results: Intriguingly, in nutrient-poor conditions UgtP levels are reduced ~ 3-fold independent of UDP-glucose. B. subtilis cells cultured under different nutrient conditions indicate that UgtP accumulation is controlled through a nutrient-dependent post-translational mechanism dependent on the Clp proteases. Notably, all three B. subtilis Clp chaperones appeared able to target UgtP for degradation during growth in nutrient-poor conditions.
Conclusions: Together these findings highlight conditional proteolysis as a mechanism for bacterial adaptation to a rapidly changing nutritional landscape.
Keywords: Cell cycle; Cell division; Cell size; ClpP; UDP-glucose; UgtP.
Conflict of interest statement
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

