Development of a New Application for Comprehensive Viability Analysis Based on Microbiome Analysis by Next-Generation Sequencing: Insights into Staphylococcal Carriage in Human Nasal Cavities
- PMID: 29625975
- PMCID: PMC5960957
- DOI: 10.1128/AEM.00517-18
Development of a New Application for Comprehensive Viability Analysis Based on Microbiome Analysis by Next-Generation Sequencing: Insights into Staphylococcal Carriage in Human Nasal Cavities
Abstract
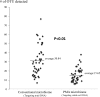
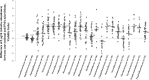
The nasal carriage rate of Staphylococcus aureus in human is 25 to 30%, and S. aureus sporadically causes severe infections. However, the mechanisms underlying staphylococcal carriage remain largely unknown. In the present study, we constructed an rpoB-based microbiome method for staphylococcal species discrimination. Based on a microbiome scheme targeting viable cell DNA using propidium monoazide (PMA) dye (PMA microbiome method), we also developed a new method to allow the comprehensive viability analysis of any bacterial taxon. To clarify the ecological distribution of staphylococci in the nasal microbiota, we applied these methods in 46 nasal specimens from healthy adults. PMA microbiome results showed that Staphylococcaceae and Corynebacteriaceae were the most predominant viable taxa (average relative abundance: 0.435262 and 0.375195, respectively), and Staphylococcus epidermidis exhibited the highest viability in the nasal microbiota. Staphylococcus aureus detection rates from nasal specimens by rpoB-based conventional and PMA microbiome methods were 84.8% (39 of 46) and 69.5% (32 of 46), respectively, which substantially exceeded the values obtained by a culture method using identical specimens (36.9%). Our results suggest that Staphylococcaceae species, especially S. epidermidis, adapted most successfully to human nasal cavity. High detection of S. aureus DNA by microbiome methods suggests that almost all healthy adults are consistently exposed to S. aureus in everyday life. Furthermore, the large difference in S. aureus detection rates between culture and microbiome methods suggests that S. aureus cells frequently exist in a viable but nonculturable state in nasal cavities. Our method and findings will contribute to a better understanding of the mechanisms underlying carriage of indigenous bacteria.IMPORTANCE Metagenomic analyses, such as 16S rRNA microbiome methods, have provided new insights in various research fields. However, conventional 16S rRNA microbiome methods do not permit taxonomic analysis of only the viable bacteria in a sample and have poor resolving power below the genus level. Our new schemes allowed for viable cell-specific analysis and species discrimination, and nasal microbiome data using these methods provided some interesting findings regarding staphylococcal nasal carriage. According to our comprehensive viability analysis, the high viability of Staphylococcus species, especially Staphylococcus epidermidis, in human nasal carriage suggests that this taxon has adapted most successfully to human nasal tissue. Also, a higher detection rate of S. aureus DNA by microbiome methods (84.8%) than by a culture method (36.9%) suggests that almost all healthy adults are consistently exposed to Staphylococcus aureus in the medium and long term. Our findings will contribute to a better understanding of the mechanisms underlying the carriage of indigenous bacteria.
Keywords: Staphylococcus; Staphylococcus aureus; metagenomics; microbial ecology.
Copyright © 2018 American Society for Microbiology.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

